Monoubiquitin carries a novel internalization signal that is appended to activated receptors - PubMed (original) (raw)
Monoubiquitin carries a novel internalization signal that is appended to activated receptors
S C Shih et al. EMBO J. 2000.
Abstract
Ubiquitin modification of signal transducing receptors at the plasma membrane is necessary for rapid receptor internalization and downregulation. We have investigated whether ubiquitylation alters a receptor cytoplasmic tail to reveal a previously masked internalization signal, or whether ubiquitin itself carries an internalization signal. Using an alpha-factor receptor-ubiquitin chimeric protein, we demonstrate that monoubiquitin can mediate internalization of an activated receptor that lacks all cytoplasmic tail sequences. Furthermore, fusion of ubiquitin in-frame to the stable plasma membrane protein Pma1p stimulates endocytosis of this protein. Ubiquitin does not carry a functional tyrosine- or di-leucine-based internalization signal. Instead, the three-dimensional structure of the folded ubiquitin polypeptide carries an internalization signal that consists of two surface patches surrounding the critical residues Phe4 and Ile44. We conclude that ubiquitin functions as a novel regulated internalization signal that can be appended to a plasma membrane protein to trigger downregulation.
Figures
Fig. 1. Cytoplasmic tail variants of Ste2p. The cytoplasmic tail of Ste2p (residues 298–431) is represented schematically. Filled circles, lysine ubiquitylation sites; open circles, lysine→arginine mutations; TMD, transmembrane domain; (g)15, Gly/Ser linker; UB, ubiquitin.
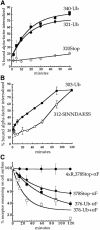
Fig. 2. Ubiquitin fused in-frame to Ste2p at amino acids 340 or 376 directs internalization of Ste2p–Ub chimeric proteins. All curves represent the average of at least three independent experiments and error bars depict the standard deviation. (A) Ste2p and Ste2p–Ub variants were introduced into ste2Δ cells and the resulting strains were assayed for their ability to internalize 35S-labeled pheromone. Cells were harvested and incubated with [35S]α–factor at 4°C, unbound α–factor was removed, and casamino acid medium pre-warmed to 30°C was added to initiate internalization. At various times aliquots of the cells were removed and the ratio of internal to total cell-associated α–factor was measured. Internalization half-times were determined by a simple exponential curve fit using Kaleidagraph software. Ste2p- 376–Ub (LHY558, •; _t_1/2 = 9.2 min); Ste2p-340–Ub (LHY844, ▴; _t_1/2 = 12.8 min); Ste2p-340Stop (LHY847, ▵; _t_1/2 = 28.3 min); Ste2p-337R,340Stop (LHY319, ⋄; _t_1/2 >200 min). (B) Wild-type (LHY558, ○), end3-1 (LHY1488, ▪), end4-1 (LHY1490, •) or pan1-20 (LHY1580, ♦) strains expressing Ste2p-376–Ub (LHP361) as their sole source of Ste2p were propagated in casamino acid medium and shifted to the non-permissive temperature of 37°C for 20 min. 35S–labeled α–factor was added and the ratio of internalized to total cell-associated α–factor was determined at different time points.
Fig. 3. Ubiquitin is sufficient to internalize Ste2p. (A and B) α–factor internalization assays were performed on cells expressing Ste2p mutants as described for Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. Internalization half-times were determined by an exponential curve fit using Kaleidagraph software, except for Ste2p-312–SINNDAKSS, which required a linear curve fit. (A) Ste2p-340–Ub (LHY844, ▪; _t_1/2 = 12.8 min); Ste2p-320Stop (LHY852, ⋄; _t_1/2 >200 min); Ste2p-321–Ub (LHY1194, •; _t_1/2 = 18.5 min). (B) Ste2p-303–Ub (LHY1212, •; _t_1/2 = 57.6 min); Ste2p-312–SINNDAKSS (LHY1078, ○; _t_1/2 = ∼76 min). The fused ubiquitin moiety in Ste2p-303–Ub and the lysine ubiquitin acceptor site in Ste2p-312–SINNDAKSS were approximately the same distance from the plasma membrane in these mutants (see Figure 1). (C) Assays that measure the number of α–factor binding sites present at the cell surface at various times after internalization at 30°C in the presence or absence of pheromone were performed on cells expressing different Ste2p variants (see Materials and methods). Ste2p-4xR,378Stop (LHY319, no α–factor, •); Ste2p-378Stop (LHY18, no α–factor, ♦); Ste2p-376–Ub (LHY558, no α–factor, ▪; + α–factor, □).
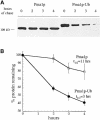
Fig. 4. Ubiquitin fused to the C-terminus of Pma1p increases its turnover rate. Strains carrying GAL-HA-PMA1 or GAL-HA-PMA1-UBI plasmids were grown to log phase in casamino acid medium containing 2% galactose and 1% raffinose to induce protein expression from the GAL1 promoter. The cells were harvested, resuspended in glucose-containing medium to inhibit Pma1p or Pma1p–Ub synthesis, and allowed to recover for 20 min at 30°C. An aliquot of cells was removed immediately (time = 0 h) and additional aliquots were removed after 2, 3 and 4 h. Cell lysates were prepared, and proteins were separated by SDS–PAGE on 6% gels and analyzed by immunoblotting with anti-HA antiserum. The immunoblots shown were developed with chemiluminescence and half-times of degrad- ation were calculated from blots of the same lysates developed with chemifluorescence to ensure that the signal was linear. (A) Degradation of Pma1p and Pma1p–Ub expressed in wild-type cells. (B) Degradation of Pma1p and Pma1–Ub turnover shown in (A) was quantified. The curves represent the average of three independent experiments and error bars represent the standard deviations.
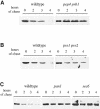
Fig. 5. Pma1p–Ub is degraded in the vacuole after internalization from the cell surface. Pma1p and Pma1p–Ub were expressed under control of the GAL1 promoter in different strains. The turnover rate of the Pma1p proteins was measured as described in Figure 4, except that cells were shifted for 20 min to the non-permissive temperature before the first aliquot of cells was withdrawn. (A) Degradation of Pma1p–Ub in pep4 prb1 cells (LHY1479, _t_1/2 = 15 h) and congenic wild-type cells (LHY1477, _t_1/2 = 2.8 h) incubated at 37°C. (B) Degradation of Pma1p–Ub in pre1 pre2 cells (LHY1473, _t_1/2 = 2.9 h) and congenic wild-type cells (LHY1471, _t_1/2 = 4.2 h) incubated at 38°C. (C) Degradation of Pma1p–Ub in pan1-20 cells (LHY1469; _t_1/2 = 13.4 h), sec6 cells (LHY1487; _t_1/2 = 11.5 h) and wild-type cells (LHY1465; _t_1/2 = 3 h) incubated at 37°C. The immunoblots shown were developed with chemiluminescence and half-times of degradation were calculated from blots of the same lysates developed with chemifluorescence to ensure that the signal was linear.
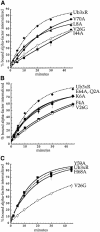
Fig. 6. Ubiquitin internalization information is not a tyrosine- or di-leucine-based signal but is carried in the three-dimensional structure of the polypeptide. (A) Sequence of ubiquitin. Shown in bold and underlined are residues that may be part of well-characterized plasma membrane protein internalization motifs. (B and C) α–factor internalization assays were performed on cells expressing different Ste2p and Ste2p–Ub mutants (see Figure 1). Cells expressing these receptors were grown to log phase in SD minimal medium and assayed as described in Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. (B) Ste2p-340–Ub (LHY713, □); Ste2p-337R,340Stop (LHY848, ⋄); Ste2p-340-DQQRLI (LHY1518, •). (C) Ste2p-376–Ub3xR (LHY1127, □); Ste2p-4xR,378Stop (LHY319, ⋄); Ste2p-376–UbV26G (LHY1157, ▪); Ste2p-376–UbL43A (LHY1162, •).
Fig. 7. Mutations in ubiquitin surface residues that inhibit Ste2p–Ub internalization. α–factor internalization assays were performed on cells expressing different Ste2p–Ub mutant receptors. The Ste2p–Ub chimeras carried mutations of ubiquitin lysines 29, 48 and 63 to arginine. Cells expressing these receptors were grown to log phase in SD minimal medium and assayed as described in Figure 2A. All curves represent the average of at least three independent experiments and error bars represent the standard deviations. (A) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbI44A (LHY1186, ▪); Ste2p-376–UbL8A (LHY1340, ▴); Ste2p-376–UbV70A (LHY1338, •). (B) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbF4A (LHY1516, ▪); Ste2p-376–UbQ2A (LHY1386, ▴); Ste2p-376–UbE64A (LHY1596, •); Ste2p-376–UbK6A (LHY1676, ▾). (C) Ste2p-376–Ub3xR (LHY1127, ♦); Ste2p-376–UbV26G (LHY1157, ⋄); Ste2p-376–UbY59A (LHY1153, ▪); Ste2p-376–UbH68A (LHY1381, •).
Fig. 8. Internalization information carried by ubiquitin is present in two surface patches bridged by a lysine residue. The three-dimensional structure of ubiquitin (Vijay-Kumar et al., 1985) is presented at two different angles in images constructed with RasMol molecular visualization software (Sayle and Milner-White, 1995). Residues essential for internalization are highlighted in magenta; surrounding residues that function in endocytosis but play a lesser role are shown in pink.
Similar articles
- A function for monoubiquitination in the internalization of a G protein-coupled receptor.
Terrell J, Shih S, Dunn R, Hicke L. Terrell J, et al. Mol Cell. 1998 Jan;1(2):193-202. doi: 10.1016/s1097-2765(00)80020-9. Mol Cell. 1998. PMID: 9659916 - Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins.
Hicke L. Hicke L. FASEB J. 1997 Dec;11(14):1215-26. doi: 10.1096/fasebj.11.14.9409540. FASEB J. 1997. PMID: 9409540 Review. - The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein.
Kölling R, Losko S. Kölling R, et al. EMBO J. 1997 May 1;16(9):2251-61. doi: 10.1093/emboj/16.9.2251. EMBO J. 1997. PMID: 9171340 Free PMC article. - Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis.
Hicke L, Riezman H. Hicke L, et al. Cell. 1996 Jan 26;84(2):277-87. doi: 10.1016/s0092-8674(00)80982-4. Cell. 1996. PMID: 8565073 - Protein regulation by monoubiquitin.
Hicke L. Hicke L. Nat Rev Mol Cell Biol. 2001 Mar;2(3):195-201. doi: 10.1038/35056583. Nat Rev Mol Cell Biol. 2001. PMID: 11265249 Review.
Cited by
- Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol.
Shamu CE, Flierman D, Ploegh HL, Rapoport TA, Chau V. Shamu CE, et al. Mol Biol Cell. 2001 Aug;12(8):2546-55. doi: 10.1091/mbc.12.8.2546. Mol Biol Cell. 2001. PMID: 11514634 Free PMC article. - STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif.
Mizuno E, Kawahata K, Kato M, Kitamura N, Komada M. Mizuno E, et al. Mol Biol Cell. 2003 Sep;14(9):3675-89. doi: 10.1091/mbc.e02-12-0823. Epub 2003 Jun 13. Mol Biol Cell. 2003. PMID: 12972556 Free PMC article. - Endocytosis of membrane receptors: two pathways are better than one.
Aguilar RC, Wendland B. Aguilar RC, et al. Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):2679-80. doi: 10.1073/pnas.0500213102. Epub 2005 Feb 14. Proc Natl Acad Sci U S A. 2005. PMID: 15710869 Free PMC article. No abstract available. - Targeted disruption of an EH-domain protein endocytic complex, Pan1-End3.
Whitworth K, Bradford MK, Camara N, Wendland B. Whitworth K, et al. Traffic. 2014 Jan;15(1):43-59. doi: 10.1111/tra.12125. Epub 2013 Oct 31. Traffic. 2014. PMID: 24118836 Free PMC article. - Epidermal growth factor cytoplasmic domain affects ErbB protein degradation by the lysosomal and ubiquitin-proteasome pathway in human cancer cells.
Glogowska A, Stetefeld J, Weber E, Ghavami S, Hoang-Vu C, Klonisch T. Glogowska A, et al. Neoplasia. 2012 May;14(5):396-409. doi: 10.1596/neo.111514. Neoplasia. 2012. PMID: 22745586 Free PMC article.
References
- Benito B., Moreno, E. and Lagunas, R. (1991) Half-life of the plasma membrane ATPase and its activating system in resting yeast cells. Biochim. Biophys. Acta, 1063, 265–268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases