A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs - PubMed (original) (raw)
A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs
P R Copeland et al. EMBO J. 2000.
Abstract
In eukaryotes, the decoding of the UGA codon as selenocysteine (Sec) requires a Sec insertion sequence (SECIS) element in the 3' untranslated region of the mRNA. We purified a SECIS binding protein, SBP2, and obtained a cDNA clone that encodes this activity. SBP2 is a novel protein containing a putative RNA binding domain found in ribosomal proteins and a yeast suppressor of translation termination. By UV cross-linking and immunoprecipitation, we show that SBP2 specifically binds selenoprotein mRNAs both in vitro and in vivo. Using (75)Se-labeled Sec-tRNA(Sec), we developed an in vitro system for analyzing Sec incorporation in which the translation of a selenoprotein mRNA was both SBP2 and SECIS element dependent. Immunodepletion of SBP2 from the lysates abolished Sec insertion, which was restored when recombinant SBP2 was added to the reaction. These results establish that SBP2 is essential for the co-translational insertion of Sec into selenoproteins. We hypothesize that the binding activity of SBP2 may be involved in preventing termination at the UGA/Sec codon.
Figures
Fig. 1. Predicted amino acid sequence of SBP2. (A) The largest ORF of the isolated SBP2 cDNA encodes a protein of 846 amino acids with a predicted molecular mass of 94.86 kDa. The positions of six peptides obtained by LC/MS analysis are underlined. Asterisks are placed above the amino acids conserved in the putative RNA binding domain. The putative nuclear localization signal is boxed. (B) BLASTp-generated alignment of SBP2 and human hypothetical protein KIAA0256 (HHP).
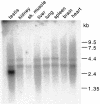
Fig. 2. Tissue distribution of SBP2 mRNA. A multiple tissue Northern blot (Clontech) of poly(A)+ RNAs from rat tissues was probed with 32P-labeled SBP2 cDNA. Molecular weight standards are indicated on the right.
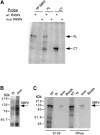
Fig. 3. UV cross-linking and immunoprecipitation of endogenous and recombinant SBP2. (A) Partially purified (PP SBP2) and recombinant Strep-tagged full-length (FL) or C–terminal (CT) SBP2 were incubated with either wild-type (wt) or mutant (mut) 32P-labeled PHGPx 3′ UTRs and subjected to UV cross-linking. After digestion with RNase A, the proteins were separated by SDS–PAGE and visualized by autoradiography. (B) Pre-immune (PI) and immune (Imm) sera raised against recombinant SBP2 were added to 1 mg of rat testicular S100 extract and complexes were precipitated with protein A–agarose. Proteins were resolved by electrophoresis, blotted to PVDF membrane and probed with anti-SBP2 peptide antiserum. (C) UV cross-linking assays were performed as described in (A) using 30 μg of S100 testicular extracts or 3 μg of partially purified SBP2 (PPure). Anti-SBP2 peptide antibody was added to the reactions as indicated, and complexes were precipitated with protein A–agarose and resolved by electrophoresis. Non-antibody-treated samples (NT) and samples treated with protein A–agarose alone (beads) were included as controls. Radiolabeled proteins were detected by PhosphorImager analysis.
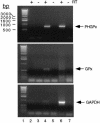
Fig. 4. Interaction of SBP2 with selenoprotein mRNAs in vivo. Pre-immune or anti-SBP2 antisera were added to cytoplasmic extracts from McArdle 7777 cells, and complexes were precipitated with protein A–agarose. The co-immunoprecipitated RNAs were extracted and used as templates for RT–PCR using PHGPx- (top), GPx- (middle) or GAPDH-specific primers (bottom). Reverse transcriptase (RT) was included as indicated. RT–PCR from immune precipitated beads is shown in lanes 4 and 5, that from pre-immune beads in lanes 2 and 3. RT–PCR from total McArdle RNA is shown in lanes 6 and 7. The PCR products were resolved by 1% agarose gel electrophoresis. Lane 1 contains a 1 kb DNA ladder.
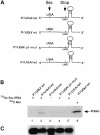
Fig. 5. Sec incorporation in vitro. (A) Diagram of the PHGPx constructs used in this study. (B) PHGPx mRNAs shown in (A) were added to reticulocyte lysates in the presence of 75Se-labeled Sec-tRNASec or [35S]Met as indicated. In vitro translated PHGPx protein was purified by BSP–agarose treatment, resolved by electrophoresis and detected by PhosphorImager analysis. 35S-labeled PHGPx is included as a control for PHGPx migration during electrophoresis (lane 6). (C) Wild-type and mutant PHGPx mRNAs were extracted at the end of in vitro translation reactions and analyzed by Northern blotting (n = 3). A representative experiment is shown.
Fig. 6. Addition of SBP2 to reticulocyte lysates. (A) Full-length (FL), N–terminal (NT) and C–terminal (CT) SBP2 mRNAs were translated in reticulocyte lysates in the presence of [35S]Met. Reactions were terminated, and proteins were resolved by SDS–PAGE. (B) PHGPx mRNAs as shown in Figure 5A were translated in reticulocyte lysates in the presence of 75Se-labeled Sec-tRNASec and in the presence or absence of the SBP2 proteins described in (A). In vitro translated PHGPx was purified on BSP–agarose and resolved by SDS–PAGE. (C) PHGPx mRNAs as indicated were translated in reticulocyte lysates as described in (B) except that all reactions were performed in the presence of pre-translated C–terminal SBP2. (D) As in (B) except that in vitro translated PHGPx protein (from P/UGU/wt construct) was blotted to PVDF and probed with anti-PHGPx antibody.
Fig. 7. SBP2 is required for Sec incorporation in reticulocyte lysates. Reticulocyte lysates were treated with either pre-immune or anti-SBP2 immune sera, and complexes were precipitated with protein A–agarose. PHGPx mRNAs as shown in Figure 5A were translated in the depleted reticulocyte lysate in the presence of 75Se-labeled Sec-tRNASec. Full-length SBP2 was added back to depleted lysates as indicated. Proteins were resolved by SDS–PAGE and detected by PhosphorImager analysis.
Similar articles
- Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2.
Bubenik JL, Miniard AC, Driscoll DM. Bubenik JL, et al. RNA Biol. 2014;11(11):1402-13. doi: 10.1080/15476286.2014.996472. RNA Biol. 2014. PMID: 25692238 Free PMC article. - Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2.
Copeland PR, Stepanik VA, Driscoll DM. Copeland PR, et al. Mol Cell Biol. 2001 Mar;21(5):1491-8. doi: 10.1128/MCB.21.5.1491-1498.2001. Mol Cell Biol. 2001. PMID: 11238886 Free PMC article. - Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes.
Chavatte L, Brown BA, Driscoll DM. Chavatte L, et al. Nat Struct Mol Biol. 2005 May;12(5):408-16. doi: 10.1038/nsmb922. Epub 2005 Apr 10. Nat Struct Mol Biol. 2005. PMID: 15821744 - Protein factors mediating selenoprotein synthesis.
Lescure A, Fagegaltier D, Carbon P, Krol A. Lescure A, et al. Curr Protein Pept Sci. 2002 Feb;3(1):143-51. doi: 10.2174/1389203023380783. Curr Protein Pept Sci. 2002. PMID: 12370018 Review. - Post-transcriptional control of selenoprotein biosynthesis.
Seeher S, Mahdi Y, Schweizer U. Seeher S, et al. Curr Protein Pept Sci. 2012 Jun;13(4):337-46. doi: 10.2174/138920312801619448. Curr Protein Pept Sci. 2012. PMID: 22708491 Review.
Cited by
- Current Understanding of Human Polymorphism in Selenoprotein Genes: A Review of Its Significance as a Risk Biomarker.
Ferreira RR, Carvalho RV, Coelho LL, Gonzaga BMS, Bonecini-Almeida MDG, Garzoni LR, Araujo-Jorge TC. Ferreira RR, et al. Int J Mol Sci. 2024 Jan 24;25(3):1402. doi: 10.3390/ijms25031402. Int J Mol Sci. 2024. PMID: 38338681 Free PMC article. Review. - Selenium Transport Mechanism via Selenoprotein P-Its Physiological Role and Related Diseases.
Saito Y. Saito Y. Front Nutr. 2021 May 28;8:685517. doi: 10.3389/fnut.2021.685517. eCollection 2021. Front Nutr. 2021. PMID: 34124127 Free PMC article. Review. - Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression.
Miniard AC, Middleton LM, Budiman ME, Gerber CA, Driscoll DM. Miniard AC, et al. Nucleic Acids Res. 2010 Aug;38(14):4807-20. doi: 10.1093/nar/gkq247. Epub 2010 Apr 12. Nucleic Acids Res. 2010. PMID: 20385601 Free PMC article. - The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function.
Papp LV, Lu J, Striebel F, Kennedy D, Holmgren A, Khanna KK. Papp LV, et al. Mol Cell Biol. 2006 Jul;26(13):4895-910. doi: 10.1128/MCB.02284-05. Mol Cell Biol. 2006. PMID: 16782878 Free PMC article. - Deciphering the role of RNA structure in translation efficiency.
Lin J, Chen Y, Zhang Y, Lin H, Ouyang Z. Lin J, et al. BMC Bioinformatics. 2022 Dec 23;23(Suppl 3):559. doi: 10.1186/s12859-022-05037-7. BMC Bioinformatics. 2022. PMID: 36564729 Free PMC article.
References
- Berry M.J., Banu, L., Chen, Y.Y., Mandel, S.J., Kieffer, J.D., Harney, J.W. and Larsen, P.R. (1991a) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature, 353, 273–276. - PubMed
- Berry M.J., Banu, L. and Larsen, P.R. (1991b) Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature, 349, 438–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 DK009878/DK/NIDDK NIH HHS/United States
- P01 HL029582/HL/NHLBI NIH HHS/United States
- F32 DK09878-01/DK/NIDDK NIH HHS/United States
- HL29582/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases