Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis - PubMed (original) (raw)
Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis
A Maor-Shoshani et al. Proc Natl Acad Sci U S A. 2000.
Abstract
When challenged by DNA-damaging agents, Escherichia coli cells respond by inducing the SOS stress response, which leads to an increase in mutation frequency by two mechanisms: translesion replication, a process that causes mutations because of misinsertion opposite the lesions, and an inducible mutator activity, which acts at undamaged sites. Here we report that DNA polymerase V (pol V; UmuC), which previously has been shown to be a lesion-bypass DNA polymerase, was highly mutagenic during in vitro gap-filling replication of a gapped plasmid carrying the cro reporter gene. This reaction required, in addition to pol V, UmuD', RecA, and single-stranded DNA (ssDNA)-binding protein. pol V produced point mutations at a frequency of 2.1 x 10(-4) per nucleotide (2.1% per cro gene), 41-fold higher than DNA polymerase III holoenzyme. The mutational spectrum of pol V was dominated by transversions (53%), which were formed at a frequency of 1.3 x 10(-4) per nucleotide (1. 1% per cro gene), 74-fold higher than with pol III holoenzyme. The prevalence of transversions and the protein requirements of this system are similar to those of in vivo untargeted mutagenesis (SOS mutator activity). This finding suggests that replication by pol V, in the presence of UmuD', RecA, and ssDNA-binding protein, is the basis of chromosomal SOS untargeted mutagenesis.
Figures
Figure 1
Preparation of the gapped plasmid. Plasmid pOC2 (A) was nicked upstream to the cro gene with restriction enzyme _Aat_II in the presence of ethidium bromide. This generated two subpopulations of nicked plasmid (B and C). Addition of exonuclease III extended the nicks into gaps in the 3′ → 5′ direction. Half of the molecules contain the cro gene in the single-stranded region.
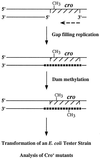
Figure 2
Outline of the cro replication fidelity assay.
Figure 3
Gap-filling DNA replication by pol V and pol III holoenzyme. Gap-filling replication was performed with pol V (MBP-UmuC) in the presence of UmuD′, RecA, and SSB, or with pol III holoenzyme, by using gapped pOC2 as a substrate. Reactions were performed in parallel in the presence and the absence of radiolabeled dTTP at 37°C for 20 min. Reaction products were fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining (Left) or phosphorimaging (Right). Based on the analysis of the phosphorimage, the amount of radiolabel incorporated into DNA by pol V was 29.4% of the amount incorporated by pol III holoenzyme. FI, supercoiled plasmid; FII, open, circular plasmid; FIII, linearized plasmid; FIV, covalently closed and relaxed plasmid; GP, gapped plasmid.
Figure 4
Spectra of mutations generated in the cro gene during in vitro replication by pol V or by pol III holoenzyme. The 201 nt of the coding region of cro are shown. Mutations generated by pol V are shown above the cro sequence, whereas mutations generated by pol III holoenzyme are shown underneath the sequence. Δ, −1 deletion; M, −2 deletion; V, +1 insertion next to the marked nucleotide. The identity of the inserted nucleotide is shown in parentheses, unless it is identical to the template nucleotide after which it was inserted. W, +2 insertion. In addition to mutations in the coding region, eight mutations were found upstream to cro: for pol V, two C → A and one each of T → G, T → A, C → T, +T; for pol III holoenzyme, C → T and G → T, one each.
Similar articles
- Activity of the purified mutagenesis proteins UmuC, UmuD', and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III.
Rajagopalan M, Lu C, Woodgate R, O'Donnell M, Goodman MF, Echols H. Rajagopalan M, et al. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10777-81. doi: 10.1073/pnas.89.22.10777. Proc Natl Acad Sci U S A. 1992. PMID: 1438275 Free PMC article. - Visualization of two binding sites for the Escherichia coli UmuD'(2)C complex (DNA pol V) on RecA-ssDNA filaments.
Frank EG, Cheng N, Do CC, Cerritelli ME, Bruck I, Goodman MF, Egelman EH, Woodgate R, Steven AC. Frank EG, et al. J Mol Biol. 2000 Mar 31;297(3):585-97. doi: 10.1006/jmbi.2000.3591. J Mol Biol. 2000. PMID: 10731413 - The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA, and SSB and is specialized for translesion replication.
Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. Reuven NB, et al. J Biol Chem. 1999 Nov 5;274(45):31763-6. doi: 10.1074/jbc.274.45.31763. J Biol Chem. 1999. PMID: 10542196 - A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V.
Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. Patel M, et al. Crit Rev Biochem Mol Biol. 2010 Jun;45(3):171-84. doi: 10.3109/10409238.2010.480968. Crit Rev Biochem Mol Biol. 2010. PMID: 20441441 Free PMC article. Review. - A Comprehensive View of Translesion Synthesis in Escherichia coli.
Fujii S, Fuchs RP. Fujii S, et al. Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
Cited by
- Reactive oxygen species accelerate de novo acquisition of antibiotic resistance in E. coli.
Qi W, Jonker MJ, de Leeuw W, Brul S, Ter Kuile BH. Qi W, et al. iScience. 2023 Oct 31;26(12):108373. doi: 10.1016/j.isci.2023.108373. eCollection 2023 Dec 15. iScience. 2023. PMID: 38025768 Free PMC article. - Comparison between complete genomes of an isolate of Pseudomonas syringae pv. actinidiae from Japan and a New Zealand isolate of the pandemic lineage.
Poulter RTM, Ho J, Handley T, Taiaroa G, Butler MI. Poulter RTM, et al. Sci Rep. 2018 Jul 19;8(1):10915. doi: 10.1038/s41598-018-29261-5. Sci Rep. 2018. PMID: 30026612 Free PMC article. - The Roles of UmuD in Regulating Mutagenesis.
Ollivierre JN, Fang J, Beuning PJ. Ollivierre JN, et al. J Nucleic Acids. 2010 Sep 30;2010:947680. doi: 10.4061/2010/947680. J Nucleic Acids. 2010. PMID: 20936072 Free PMC article. - Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I.
Camps M, Naukkarinen J, Johnson BP, Loeb LA. Camps M, et al. Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9727-32. doi: 10.1073/pnas.1333928100. Epub 2003 Aug 8. Proc Natl Acad Sci U S A. 2003. PMID: 12909725 Free PMC article. - Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells.
Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Kannouche P, et al. Genes Dev. 2001 Jan 15;15(2):158-72. doi: 10.1101/gad.187501. Genes Dev. 2001. PMID: 11157773 Free PMC article.
References
- Little J W, Mount D W. Cell. 1982;29:11–22. - PubMed
- Sassanfar M, Roberts J F. J Mol Biol. 1990;212:79–96. - PubMed
- Eggleston A K, West S C. Trends Genet. 1996;12:20–26. - PubMed
- Cox M M. Genes Cells. 1998;3:65–78. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous