Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli - PubMed (original) (raw)
Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli
K Walker et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The cDNA clone for a 10-deacetylbaccatin III-10-O-acetyl transferase, which catalyzes formation of the last diterpene intermediate in the Taxol biosynthetic pathway, has been isolated from Taxus cuspidata. By using consensus sequences from an assembly of transacylases of plant origin and from many deduced proteins of unknown function, a homology-based PCR cloning strategy was employed to amplify initially a 911-bp gene fragment of the putative taxane C-10 hydroxyl acetyl transferase from Taxus. This amplicon was used to screen a cDNA library constructed from mRNA isolated from methyl jasmonate-induced Taxus cells, from which the full-length 10-deacetylbaccatin III-10-O-transacetylase sequence was obtained. Expression of the ORF from pCWori(+) in Escherichia coli JM109 afforded a functional enzyme, as determined by (1)H-NMR and MS verification of the product baccatin III derived from 10-deacetylbaccatin III and acetyl CoA. The full-length cDNA has an ORF of 1,320 bp corresponding to a deduced protein of 440 residues with a calculated molecular weight of 49,052, consistent with the size of the operationally soluble, monomeric, native acetyl transferase. The recombinant acetyl transferase has a pH optimum of 7.5, has K(m) values of 10 microM and 8 microM for 10-deacetylbaccatin III and acetyl CoA, respectively, and is apparently regiospecific toward the 10-hydroxyl group of the taxane ring. Amino acid sequence comparison of 10-deacetylbaccatin III-10-O-acetyl transferase with taxadienol-5-O-acetyl transferase and with other known acyl transferases of plant origin indicates a significant degree of similarity between these enzymes (80% and 64-67%, respectively).
Figures
Figure 1
(A) Outline of the Taxol biosynthetic pathway. The cyclization of geranylgeranyl diphosphate to taxadiene by taxadiene synthase and the hydroxylation to taxadien-5α-ol by taxadiene 5α-hydroxylase (a), the acetylation of taxadien-5α-ol by taxa-4(20),11(12)-dien-5α-ol-_O_-acetyl transferase (b), the conversion of 10-deacetylbaccatin III (10-DAB) to baccatin III by 10-deacetylbaccatin III-10-_O_-acetyl transferase (DBAT) (c), and the side-chain attachment to baccatin III to form Taxol (d) are highlighted. The broken arrow indicates several as yet undefined steps. (B) Postulated biosynthetic scheme for the formation of the oxetane, present in Taxol and related late-stage taxoids, in which the 4(20)-ene-5α-ol is converted to the 4(20)-ene-5α-yl acetate, followed by epoxidation to the 4(20)-epoxy-5α-acetoxy group and then intramolecular rearrangement to the 4-acetoxy oxetane moiety. Ac, acetyl; Bz, benzoyl; _t_-BOC, tertiary-butoxycarbonyl; OPP, diphosphate.
Figure 2
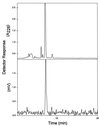
Radio-HPLC analysis of the biosynthetic product (retention time = 7.0 ± 0.1 min) generated from 10-DAB and [2-3H]acetyl CoA by the recombinant acetyl transferase. The UV profile (Upper) and the coincident radioactivity profile (Lower) coincide with the retention time of authentic baccatin III. For column, solvent, flow rates, and counting details, see Materials and Methods.
Figure 3
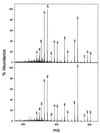
Combined reverse-phase HPLC-atmospheric pressure chemical ionization MS analysis of the biosynthetic product (retention time = 8.6 ± 0.1 min) generated by the recombinant acetyl transferase with 10-DAB and acetyl CoA as cosubstrates (Upper spectrum) and of authentic baccatin III (retention time = 8.6 ± 0.1 min;Lower spectrum). The diagnostic mass spectral fragment ions are at m/z 605 (P + NH4+), 587 (PH+), 572 (PH+ − CH3), 527 (PH+ − CH3COOH), and 509 (PH+ − (CH3COOH − H2O). For column, solvent, flow rates, and instrument details, see Materials and Methods.
Figure 4
Deduced amino acid sequence comparison of DBAT (accession no. AF193765) from T. cuspidata, taxadien-5α-ol-_O_-acetyl transferase (TAT, accession no. AF190130) from T. cuspidata, benzylalcohol acetyl transferase (BEAT, accession no. AF043464) from Clarkia breweri, and deacetylvindoline-4-_O_-acetyl transferase (DAT, accession no. AF053307) from Catharanthus roseus. Residues boxed in black indicate positional identity for at least two of the compared sequences; similar amino acids are indicated by gray shading. Asterisks (*) indicate conserved residues in all acetyl transferase sequences of plant origin. Pound signs (#) indicate a putative acyl group transfer motif (HXXXDG) present in three of the four sequences. The alignment was created with the
pileup
program (Wisconsin Package Version 9.0; Genetics Computer Group, Madison, WI).
Similar articles
- Molecular cloning of a taxa-4(20),11(12)-dien-5alpha-ol-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli.
Walker K, Schoendorf A, Croteau R. Walker K, et al. Arch Biochem Biophys. 2000 Feb 15;374(2):371-80. doi: 10.1006/abbi.1999.1609. Arch Biochem Biophys. 2000. PMID: 10666320 - Partial purification and characterization of acetyl coenzyme A: taxa-4(20),11(12)-dien-5alpha-ol O-acetyl transferase that catalyzes the first acylation step of taxol biosynthesis.
Walker K, Ketchum RE, Hezari M, Gatfield D, Goleniowski M, Barthol A, Croteau R. Walker K, et al. Arch Biochem Biophys. 1999 Apr 15;364(2):273-9. doi: 10.1006/abbi.1999.1125. Arch Biochem Biophys. 1999. PMID: 10190984 - Molecular cloning and heterologous expression of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus x media.
Guo B, Kai G, Gong Y, Jin H, Wang Y, Miao Z, Sun X, Tang K. Guo B, et al. Mol Biol Rep. 2007 Jun;34(2):89-95. doi: 10.1007/s11033-006-9018-6. Epub 2006 Nov 9. Mol Biol Rep. 2007. PMID: 17094009 - The taxol pathway 10-O-acetyltransferase shows regioselective promiscuity with the oxetane hydroxyl of 4-deacetyltaxanes.
Ondari ME, Walker KD. Ondari ME, et al. J Am Chem Soc. 2008 Dec 17;130(50):17187-94. doi: 10.1021/ja8067534. J Am Chem Soc. 2008. PMID: 19007225 - Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2alpha-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli.
Walker K, Croteau R. Walker K, et al. Proc Natl Acad Sci U S A. 2000 Dec 5;97(25):13591-6. doi: 10.1073/pnas.250491997. Proc Natl Acad Sci U S A. 2000. PMID: 11095755 Free PMC article.
Cited by
- Synthesis and In Vitro Evaluation of Taxol oxetane ring D precursors.
Kaspera R, Cape JL, Faraldos JA, Ketchum RE, Croteau RB. Kaspera R, et al. Tetrahedron Lett. 2010 Apr 14;51(15):2017-2019. doi: 10.1016/j.tetlet.2010.02.033. Tetrahedron Lett. 2010. PMID: 20305723 Free PMC article. - Molecular evolution of paclitaxel biosynthetic genes TS and DBAT of Taxus species.
Hao da C, Yang L, Huang B. Hao da C, et al. Genetica. 2009 Mar;135(2):123-35. doi: 10.1007/s10709-008-9257-7. Epub 2008 Mar 8. Genetica. 2009. PMID: 18327645 - Seasonal-based temporal changes fluctuate expression patterns of TXS, DBAT, BAPT and DBTNBT genes alongside production of associated taxanes in Taxus baccata.
Nasiri J, Naghavi MR, Alizadeh H, Moghadam MR. Nasiri J, et al. Plant Cell Rep. 2016 May;35(5):1103-19. doi: 10.1007/s00299-016-1941-y. Epub 2016 Feb 16. Plant Cell Rep. 2016. PMID: 26883228 - Salicylic Acid-Responsive Factor TcWRKY33 Positively Regulates Taxol Biosynthesis in Taxus chinensis in Direct and Indirect Ways.
Chen Y, Zhang H, Zhang M, Zhang W, Ou Z, Peng Z, Fu C, Zhao C, Yu L. Chen Y, et al. Front Plant Sci. 2021 Aug 9;12:697476. doi: 10.3389/fpls.2021.697476. eCollection 2021. Front Plant Sci. 2021. PMID: 34434205 Free PMC article. - Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells.
Lenka SK, Boutaoui N, Paulose B, Vongpaseuth K, Normanly J, Roberts SC, Walker EL. Lenka SK, et al. BMC Genomics. 2012 Apr 24;13:148. doi: 10.1186/1471-2164-13-148. BMC Genomics. 2012. PMID: 22530557 Free PMC article.
References
- Croteau R, Hefner J, Hezari M, Lewis N G. In: Phytochemicals and Health. Flores H E, Gustine D L, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1995. pp. 94–104.
- Floss H G, Mocek U. In: Taxol: Science and Applications. Suffness M, editor. Boca Raton, FL: CRC; 1995. pp. 191–208.
- Walker K, Ketchum R E B, Hezari M, Gatfield D, Golenowski M, Barthol A, Croteau R. Arch Biochem Biophys. 1999;364:273–279. - PubMed
- Walker, K., Schoendorf, A. & Croteau, R. (2000) Arch. Biochem. Biophys., in press. - PubMed
- Hezari M, Croteau R. Planta Med. 1997;63:291–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases