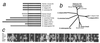Heterochromatic deposition of centromeric histone H3-like proteins - PubMed (original) (raw)
Heterochromatic deposition of centromeric histone H3-like proteins
S Henikoff et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Centromeres of most organisms are embedded within constitutive heterochromatin, the condensed regions of chromosomes that account for a large fraction of complex genomes. The functional significance of this centromere-heterochromatin relationship, if any, is unknown. One possibility is that heterochromatin provides a suitable environment for assembly of centromere components, such as special centromeric nucleosomes that contain distinctive histone H3-like proteins. We describe a Drosophila H3-like protein, Cid (for centromere identifier) that localizes exclusively to fly centromeres. When the cid upstream region drives expression of H3 and H2B histone-green fluorescent protein fusion genes in Drosophila cells, euchromatin-specific deposition results. Remarkably, when the cid upstream region drives expression of yeast, worm, and human centromeric histone-green fluorescent protein fusion proteins, localization is preferentially within Drosophila pericentric heterochromatin. Heterochromatin-specific localization also was seen for yeast and worm centromeric proteins constitutively expressed in human cells. Preferential localization to heterochromatin in heterologous systems is unexpected if centromere-specific or site-specific factors determine H3-like protein localization to centromeres. Rather, the heterochromatic state itself may help localize centromeric components.
Figures
Figure 1
Comparative sequence analysis of histone H3-like proteins used in this study. (a) Cartoon depicting nucleosomal core regions and unaligned N-terminal tails. Boxes representing the N-terminal tails are shaded differentially to depict the absence of sequence similarity between H3-like proteins. (b) Phylogenetic tree for the core region. Proteins used in this study are underlined. (c) Boxshade multiple alignment for the core region of the proteins used in this study.
Figure 2
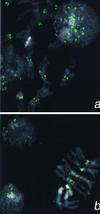
Detection of a Cid epitope at centromeric constrictions and in an interphase nucleus by using anti-Cid antiserum (1:5,000). (a) Kc167 cells. (b) Larval neuroblasts. Cid signal is shown in green 4′,6-diamidino-2-phenylindole staining in gray.
Figure 3
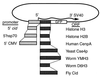
Plasmid constructs used for transient transfection. In each case, the full protein-coding sequence is fused to a 6-aa spacer followed by the N terminus of GFP (27).
Figure 4
Localization of H3-like protein-GFP fusions to centromeres of Drosophila. Kc167 cells were transfected with GFP fusion constructs driven by the cid promoter (a and d–k) or by basal (b) or induced (c) expression from the Hsp70 promoter. (a) Localization of Cid-GFP to the centromeres in a metaphase spread and to intense foci in interphase nuclei. Similar foci are seen in many interphase nuclei when Cid-GFP is expressed from the Hsp70 promoter (b and c) and are 10-fold more intense (185 ± 61 vs. 2,085 ± 979 pixel intensity) when this promoter has been induced (c). (d) Euchromatic labeling by _cid_-driven H3-GFP. Localization of GFP fusions on representative X chromosomes from mitotic figures with Drosophila H3 (e), Drosophila H2B (f), Drosophila Cid (g), human CENP-A (h), S. cerevisiae Cse4p (i), C. elegans HCP-3 (SWISS-PROT YMH3_CAEEL) (j), and C. elegans D6H3 (k) (GenBank accession no. AAB37052). GFP fluorescence was consistently lower with the H3-like heterolog-GFP fusion proteins than with the Cid- or the histone-GFP fusions. The X centromere is indicated with an arrowhead. The X chromosome is acrocentric, and the heterochromatin coheres well in metaphase spreads; thus, the position of the centromere can be identified by morphology alone. Identification of centromeres in metaphase spreads was confirmed by detection of the POLO kinetochore epitope. GFP signal is shown in green, the POLO epitope is shown in red (a, d, e, and g), and 4′,6-diamidino-2-phenylindole staining is shown in gray. Coincidence of GFP and POLO is yellow.
Figure 5
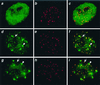
Colocalization of H3-like proteins with centromeres in human interphase nuclei. HeLa cells were transfected with GFP fusions of D. melanogaster H3 (a–c), C. elegans HCP-3 (d–f), or S. cerevisiae Cse4p (g–i) driven by the constitutive cytomegalovirus promoter. Cells were fixed and stained with ACA serum against centromeres (red); the GFP signal is shown in green. Representative interphase nuclei are shown. We chose the 60 brightest GFP spots in d and g; 70% of these spots colocalized with the ≈60 ACA spots (f and i). Arrows point to representative small spots and arrowheads point to large spots.
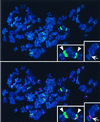
Figure 6
Colocalization of HCP-3-GFP with heterochromatin in human metaphase chromosomes. See legend to Fig. 5. 4′,6-Diamidino-2-phenylindole staining of DNA is shown in blue. Insets show two chromosomes with strong pericentric HCP-3-GFP signals (Left) and one chromosome with centromeric as well as telomeric HCP-3-GFP localization (Right).
Similar articles
- Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation.
Yang J, Sun S, Zhang S, Gonzalez M, Dong Q, Chi Z, Chen YH, Li F. Yang J, et al. PLoS Genet. 2018 Aug 8;14(8):e1007572. doi: 10.1371/journal.pgen.1007572. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30089114 Free PMC article. - Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications.
Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Nakagawa H, et al. Genes Dev. 2002 Jul 15;16(14):1766-78. doi: 10.1101/gad.997702. Genes Dev. 2002. PMID: 12130537 Free PMC article. - A role of the Trx-G complex in Cid/CENP-A deposition at Drosophila melanogaster centromeres.
Piacentini L, Marchetti M, Bucciarelli E, Casale AM, Cappucci U, Bonifazi P, Renda F, Fanti L. Piacentini L, et al. Chromosoma. 2019 Dec;128(4):503-520. doi: 10.1007/s00412-019-00711-x. Epub 2019 Jun 16. Chromosoma. 2019. PMID: 31203392 - Centromeres and variant histones: what, where, when and why?
Smith MM. Smith MM. Curr Opin Cell Biol. 2002 Jun;14(3):279-85. doi: 10.1016/s0955-0674(02)00331-9. Curr Opin Cell Biol. 2002. PMID: 12067649 Review. - Centromeric heterochromatin: the primordial segregation machine.
Bloom KS. Bloom KS. Annu Rev Genet. 2014;48:457-84. doi: 10.1146/annurev-genet-120213-092033. Epub 2014 Sep 18. Annu Rev Genet. 2014. PMID: 25251850 Free PMC article. Review.
Cited by
- Localization of Drosophila CENP-A to non-centromeric sites depends on the NuRD complex.
Demirdizen E, Spiller-Becker M, Förtsch A, Wilhelm A, Corless S, Bade D, Bergner A, Hessling B, Erhardt S. Demirdizen E, et al. Nucleic Acids Res. 2019 Dec 16;47(22):11589-11608. doi: 10.1093/nar/gkz962. Nucleic Acids Res. 2019. PMID: 31713634 Free PMC article. - De novo kinetochore assembly requires the centromeric histone H3 variant.
Collins KA, Castillo AR, Tatsutani SY, Biggins S. Collins KA, et al. Mol Biol Cell. 2005 Dec;16(12):5649-60. doi: 10.1091/mbc.e05-08-0771. Epub 2005 Oct 5. Mol Biol Cell. 2005. PMID: 16207811 Free PMC article. - Dual recognition of CENP-A nucleosomes is required for centromere assembly.
Carroll CW, Milks KJ, Straight AF. Carroll CW, et al. J Cell Biol. 2010 Jun 28;189(7):1143-55. doi: 10.1083/jcb.201001013. Epub 2010 Jun 21. J Cell Biol. 2010. PMID: 20566683 Free PMC article. - Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins.
Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJ, van der Zaal BJ. Lindhout BI, et al. Nucleic Acids Res. 2007;35(16):e107. doi: 10.1093/nar/gkm618. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704126 Free PMC article. - CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells.
Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. Ando S, et al. Mol Cell Biol. 2002 Apr;22(7):2229-41. doi: 10.1128/MCB.22.7.2229-2241.2002. Mol Cell Biol. 2002. PMID: 11884609 Free PMC article.
References
- Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig, Germany: F. C. W. Vogel; 1882.
- Wiens G R, Sorger P K. Cell. 1998;93:313–316. - PubMed
- Csink A K, Henikoff S. Trends Genet. 1998;14:200–204. - PubMed
- Willard H F. Curr Op Genet Dev. 1998;8:219–225. - PubMed
- Murphy T D, Karpen G H. Cell. 1998;93:317–320. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous