Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion - PubMed (original) (raw)
Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion
P Steinberger et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Studies of naturally occurring polymorphisms of the CCR5 gene have shown that deletion of the functional receptor or reduced expression of the gene can have beneficial effects in preventing HIV-1 infection or delaying disease. Because these polymorphisms are found in otherwise healthy people, strategies that aim to prevent or limit expression of CCR5 should be beneficial in the treatment of HIV-1 disease. To test this approach we have developed a CCR5-specific single-chain antibody that was expressed intracellularly and retained in the endoplasmic reticulum. This CCR5-intrabody efficiently blocked surface expression of human and rhesus CCR5 and thus prevented cellular interactions with CCR5-dependent HIV-1 and simian immunodeficiency virus envelope glycoprotein. Intrabody-expressing cells were shown to be highly refractory to challenge with R5 HIV-1 viruses or infected cells. These results suggest that gene therapy approaches that deliver this intracellular antibody could be of benefit to infected individuals. Because the antibody reacts with a conserved primate epitope on CCR5 this strategy can be tested in nonhuman lentivirus models of HIV-1 disease.
Figures
Figure 1
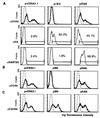
Inhibition of transient CCR5 surface expression upon cotransfection with ST6 intrabody-encoding plasmid, pIB6. (A) 293T cells were transiently transfected with plasmid encoding human CCR5 and were cotransfected with the indicated plasmids: control plasmid, pcDNA3.1/Zeo, plasmid encoding ST6, pIB6, or with plasmid encoding RANTES, pRAN. (Top) Cell surface staining using a CCR5-specific antibody (clone 5C7, bold lines) or with an irrelevant mouse antibody for control purposes (dashed lines). (Middle) Staining of permeabilized cells with an HA-tag-specific antibody to detect intrabody or intrakine. (Bottom) Staining of permeabilized cells for intracellular RANTES expression with an anti-human RANTES antibody. (B) 293T cells were transiently transfected with plasmid encoding rhesus CCR5 and were cotransfected with plasmids as indicated. Cells were surface stained for CCR5 expression as described above. (C) 293T cells were transiently transfected with plasmid encoding human CXCR4 and were cotransfected with plasmids as indicated. Transfected cells were surface stained with a CXCR4-specific antibody (bold lines), or control stained with an irrelevant mouse antibody (dashed lines).
Figure 2
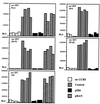
Cotransfection of ST6-encoding plasmid (pIB6) inhibits CCR5-dependent cell fusion between target cells transiently transfected to express CCR5 and CD4 and effector cells expressing env derived from various HIV-1 or SIV strains. For determination of background luciferase activity, a set of target cells was cotransfected with plasmids encoding CD4 and luciferase but no CCR5-encoding plasmid (“no CCR5”). “Control” indicates fusion assays using target cells cotransfected with plasmids encoding CCR5, CD4, luciferase, and control plasmid pcDNA3.1/Zeo containing no insert. “pIB6” indicates luciferase activity where target cells were cotransfected with plasmids encoding CCR5, CD4, luciferase, and ST6. “pRAN” indicates luciferase activity, where target cells were cotransfected with plasmids encoding CCR5, CD4, luciferase, and RANTES. Relative light units (RLU) are shown on the _y_-axis. Each bar represents luciferase activity from one well of target cells.
Figure 3
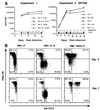
Analysis of untransduced PM1 cells (PM1-P) and PM1 cell lines transduced to express ST6 intrabody (PM1–6) or control intrabody (PM1-RAI3) and a cell clone derived from the cell line PM1-RAI3 (PM1-RAI3–5) by flow cytometry. (A) PM1 cells were stained with CCR5-specific antibody (bold line), CD4-specific antibody (dashed line), or irrelevant negative control antibody (thin line). (B) PM1 cells were permeabilized and stained with an HA-tag-specific antibody. Percentages are number of cells staining positive. (C) Analysis of unpermeablized PM1 cells for cell surface scFv. PM1 cells were stained with an HA-tag-specific antibody.
Figure 4
Cell–cell fusion assay using parental PM1 cells (PM1-P) and clones derived from the control transduced PM1 cells (PM1-RAI3–5) and from PM1 cells transduced with ST6-encoding retrovirus (PM1–6-G). PM1 cells were infected with a vaccinia recombinant encoding T7 RNA polymerase. To assess background luciferase activity, parental PM1 cells were infected with vaccinia virus encoding β-galactosidase instead (PM1-P-lacZ). Reporter gene activity upon coculturing of PM1 cells with 293T cells expressing HIV-1 env and transfected with reporter plasmid is shown as relative light units (RLU) on the _y_-axis. Pairs of individual experiments are shown.
Figure 5
ST6 intrabody-expressing PM1 cells are resistant to challenge by R5 HIV-1 viruses and infected cells. (A) ST6 intrabody-expressing PM1 cells are resistant to cell-free infection with the R5 HIV-1 isolates SF162 and JR-CSF. ST6-expressing PM1 cell lines and clones (dashed lines), parental PM1 cells, and transduced control lines (solid lines) were infected and monitored for p24 levels at indicated times. In experiment 1 the parental PM1 cell line (PM1-P) and the ST6-expressing PM1–6 line were infected with the isolates SF162 and JR-CSF. In experiment 2, HIV-1 SF162 was used to infect PM1 cell lines and clones as indicated. (B) ST6 intrabody-expressing PM1 cells are resistant to cell-mediated R5 HIV-1 infection. PM1 cell cultures infected with the R5-HIV-1 reporter virus NFN-SX-r-HSAS were cocultured with the parental PM1 cell line (PM1-P) and the transduced PM1 clones PM1–6-G and PM1-RAI3–5. Double staining of cells for intrabody expression (HA-FITC) and reporter virus infection (HSA-PE) after 3 and 7 days of coculturing is shown.
Similar articles
- T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery.
Swan CH, Bühler B, Steinberger P, Tschan MP, Barbas CF 3rd, Torbett BE. Swan CH, et al. Gene Ther. 2006 Oct;13(20):1480-92. doi: 10.1038/sj.gt.3302801. Epub 2006 Jun 1. Gene Ther. 2006. PMID: 16738691 - Bovine alpha-2-HS-glycoprotein functions as a booster antigen for efficiently stimulating humoral immune responses to CCR5 and SIVmac239 envelope glycoprotein.
Otsubo Y, Yashiro S, Nozaki K, Matsuura K, Kiyonaga K, Mitsumata R, Takahashi Y, Masuyama M, Muneoka A, Takamune N, Shoji S, Misumi S. Otsubo Y, et al. Biochem Biophys Res Commun. 2014 Jan 3;443(1):301-7. doi: 10.1016/j.bbrc.2013.11.098. Epub 2013 Dec 2. Biochem Biophys Res Commun. 2014. PMID: 24309114 - Host and Viral Factors in HIV-Mediated Bystander Apoptosis.
Garg H, Joshi A. Garg H, et al. Viruses. 2017 Aug 22;9(8):237. doi: 10.3390/v9080237. Viruses. 2017. PMID: 28829402 Free PMC article. Review.
Cited by
- Stem cell-based anti-HIV gene therapy.
Kitchen SG, Shimizu S, An DS. Kitchen SG, et al. Virology. 2011 Mar 15;411(2):260-72. doi: 10.1016/j.virol.2010.12.039. Epub 2011 Jan 17. Virology. 2011. PMID: 21247612 Free PMC article. Review. - A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model.
Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, Kittipongdaja P, Chen A, Bristol G, Galic Z, Zack JA, Yang O, Chen IS, Lee B, An DS. Shimizu S, et al. Blood. 2010 Feb 25;115(8):1534-44. doi: 10.1182/blood-2009-04-215855. Epub 2009 Dec 17. Blood. 2010. PMID: 20018916 Free PMC article. - Antibodies inside of a cell can change its outside: Can intrabodies provide a new therapeutic paradigm?
Marschall AL, Dübel S. Marschall AL, et al. Comput Struct Biotechnol J. 2016 Jul 31;14:304-8. doi: 10.1016/j.csbj.2016.07.003. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27570612 Free PMC article. Review. - The production and application of single-chain antibody fragments.
Blazek D, Celer V. Blazek D, et al. Folia Microbiol (Praha). 2003;48(5):687-98. doi: 10.1007/BF02993480. Folia Microbiol (Praha). 2003. PMID: 14976730 Review. - Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases.
Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Perez EE, et al. Nat Biotechnol. 2008 Jul;26(7):808-16. doi: 10.1038/nbt1410. Epub 2008 Jun 29. Nat Biotechnol. 2008. PMID: 18587387 Free PMC article.
References
- Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. Nature (London) 1998;391:240. - PubMed
- Berger E A, Murphy P M, Farber J M. Annu Rev Immunol. 1999;17:657–700. - PubMed
- Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. - PubMed
- McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. Lancet. 1998;352:866–870. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical