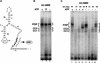ATP can be dispensable for prespliceosome formation in yeast - PubMed (original) (raw)
. 2000 Jan 1;14(1):97-107.
Affiliations
- PMID: 10640279
- PMCID: PMC316341
ATP can be dispensable for prespliceosome formation in yeast
R Perriman et al. Genes Dev. 2000.
Abstract
The first ATP-dependent step in pre-mRNA splicing involves the stable binding of U2 snRNP to form the prespliceosome. We show that a prespliceosome-like complex forms in the absence of ATP in yeast extracts lacking the U2 suppressor protein CUS2. These complexes display the same pre-mRNA and U snRNA requirements as authentic prespliceosomes and can be chased through the splicing pathway, indicating that they are a functional intermediate in the spliceosome assembly pathway. ATP-independent prespliceosome-like complexes are also observed in extracts containing a mutant U2 snRNA. Loss of CUS2 does not bypass the role of PRP5, an RNA helicase family member required for ATP-dependent prespliceosome formation. Genetic interactions between CUS2 and a heat-sensitive prp5 allele parallel those observed between CUS2 and U2, and suggest that CUS2 mediates functional interactions between U2 RNA and PRP5. We propose that CUS2 enforces ATP dependence during formation of the prespliceosome by brokering an interaction between PRP5 and the U2 snRNP that depends on correct U2 RNA structure.
Figures
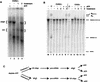
Figure 1
Splicing extracts from an ATP-depleted CUS2Δ yeast strain produce a novel complex on actin (A) and RP51A (B–D) pre-mRNA. Native gel analysis of spliceosome assembly for (A) CUS2+ (lanes 1–3) or CUS2Δ (lanes 4–6) yeast strains. (+; Lanes 1,4) +2 m
m
ATP; (P; lanes 3,6) 2 m
m
AMP–PCP; (-; lanes 2,5) no addition. Brackets: (CC) commitment complexes; (P/SP) prespliceosomes/spliceosomes. (B) Wild-type or mutant pre-RP51A substrates. (Lanes 1–4) ATP-depleted CUS2+; (WT; lanes 2,5) CUS2Δ splicing extracts incubated with wild-type RP51A; (5′; lanes 3,6) 5′ splice site mutant; (BP; lanes 4,7) branchpoint mutant pre-mRNA. Lane 1 shows P/SP formation on WT RP51A pre-mRNA in CUS2+ extracts with 2 m
m
ATP. Brackets are as for A, except that commitment complexes for pre-RP51A form two migrating species (CC1 and CC2). (C) CUS2Δ extracts treated with oligonucleotides (10) to U1 (lanes 2,6), U2 (lanes 3,7), and U6 snRNAs (lanes 4,8). Lanes 1_–_4 have 2 m
m
ATP added with pre-RP51A substrates after oligonucleotide-ablation; lanes 5–8 have no ATP. Brackets are as for B.
Figure 2
The novel ATP-independent complex can chase to spliceosomes. (A) The complex is resistant to U2 oligonucleotide. Native gel analysis of complexes formed with no U2 oligonucleotide (lanes 1,2), 45 n
m
U2 oligonucleotide before (lane 3) or after (lane 4) adding pre-actin. Lane 1 has 2 m
m
ATP. Bracketed species as for Fig. 1A. (B) Analysis of pre-actin RNA-spliced products after treatments 1 or 2. Lanes 2–6 are splicing reactions done in ATP-depleted, CUS2Δ extracts. Lanes 7–12 are identical splicing reactions done in CUS2+ extracts. Lanes 1 and 7 (C) contain 2 m
m
ATP and preactin, but no U2 oligonucleotide. Lane M is untreated preactin. Splice products from top to bottom of gel are as follows: lariat–3′ exon; excised lariat; precursor preactin; mature actin mRNA; released 5′ exon. (C) Schematic representation of two treatments for chasing the ATP-independent prespliceosome to spliceosomes.
Figure 3
(A) rCUS2 addition disrupts formation of the novel ATP-independent complex. Addition of protein dilution buffer (B, lane 2), 50, 100, or 350 n
m
rCUS2 (lanes 3–5), or rY48D (lanes 6_–_8). Lane 1 has 2 m
m
ATP and protein dilution buffer. Brackets are as for Fig. 2. (B) The rate of formation of ATP-independent prespliceosomes is reduced compared with ATP-dependent prespliceosomes. Analysis of complexes formed on pre-RP51A after addition to U6-depleted CUS2Δ (lanes 1–10) or CUS2+ (lanes 11_–_15) splicing extracts. Lanes 1–5 are ATP depleted, lanes 6–15 contain 2 m
m
ATP. Samples were taken at 0 (lanes 1,6,11), 1 (lanes 2,7,12), 5 (lanes 3,8,13), 20 (lanes 4,9,14), or 40 (lanes 5,10,15) min after addition of pre-RP51A. Brackets are as for Fig. 2.
Figure 4
(A) CUS2Δ cannot bypass the requirement for Prp5 in prespliceosome assembly. Splicing extracts isolated from prp5-1 (Ruby et al. 1993) yeast strains containing CUS2 (lanes 1,4) or genetically depleted for CUS2 (lanes 2,5) were incubated at 25°C (lanes 1–3) or 37°C (lanes 4–6) prior to addition of pre-RP51A. Lanes 3 and 6 contain CUS2Δ splicing extracts containing wild-type PRP5 preincubated at 25°C (lane 3) or 37°C (lanes 6). Brackets are as for Fig. 3. (B) CUS2 and PRP5 show a genetic interaction. Analysis of growth at 25°C of strains containing wild-type or one of prp5-1 or prp5-3 temperature-sensitive mutations of PRP5 in combination with wild type or CUS2Δ. (+++) Wild-type growth; (++) intermediate growth; (+) slow growth; (−) dead. (C) CUS2-9 and CUS2-25 suppressor proteins can suppress temperature sensitivity of the prp5-1 allele. Growth of SRY5-1 with URA+ plasmids expressing one of wild-type, or CUS2 suppressor proteins as indicated. The plate was incubated at 36°C for 4 days.
Figure 5
Splicing extracts from yeast strains containing CUS2 and mutant U2 allele, U2-QMB′ also form prespliceosomes without ATP. (A) Secondary structure fold of a 5′ portion (from nucleotides 48–120) of yeast U2 snRNA showing base substitutions in U2-QMB′. The essential stem–loop IIa and conserved structural feature, stem IIb are indicated. The phylogenetically conserved downstream region is in boldface. (B) Native gel analysis of complex formation on pre-RP51A in ATP-depleted splicing extracts from CUS2+, U2-QMB′ yeast strains. (Lane 1) 2 m
m
ATP; (lane 2) 2 m
m
AMP–PCP; (lane 3) no additions. Bracketed species are as for Figs. 1 and 2. (C) Native gel analysis of U snRNA requirements for ATP-independent prespliceosomes in splicing extracts from U2-QMB′ expressing yeast. Extracts were treated with oligonucleotides designed toanneal to U1 (lanes 2,6), U2 (lanes 3,7), and U6 snRNAs (lanes 4,8). (Lanes 1–4) 2 m
m
ATP with pre-RP51A substrates after the oligonucleotide-ablation; (lanes 5–8) no ATP. Brackets are as for B.
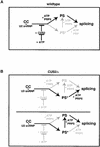
Figure 6
A working model for the role of CUS2 in regulating the ATP dependence of U2 snRNP recruitment and prespliceosome assembly. (A) In the presence of CUS2, U2 snRNP recruitment and prespliceosome formation cannot proceed without the addition of ATP (black pathway). (B) Without CUS2 protein, the U2 snRNP can enter the splicing pathway without ATP to form the functional intermediate prespliceosome-like complex (PS*). Because we cannot distinguish between PS* being on the same or alternate splicing pathways, both possibilities are depicted. As for A, the black pathway represents each alternative. Loss of CUS2 does not bypass the requirement for PRP5 and ATP in the formation of ATP-dependent prespliceosomes, hence, both pathways in B show steps beyond PS* requiring both PRP5 and ATP.
Similar articles
- ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA.
Perriman R, Barta I, Voeltz GK, Abelson J, Ares M Jr. Perriman R, et al. Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):13857-62. doi: 10.1073/pnas.2036312100. Epub 2003 Nov 10. Proc Natl Acad Sci U S A. 2003. PMID: 14610285 Free PMC article. - Cus2 enforces the first ATP-dependent step of splicing by binding to yeast SF3b1 through a UHM-ULM interaction.
Talkish J, Igel H, Hunter O, Horner SW, Jeffery NN, Leach JR, Jenkins JL, Kielkopf CL, Ares M. Talkish J, et al. RNA. 2019 Aug;25(8):1020-1037. doi: 10.1261/rna.070649.119. Epub 2019 May 20. RNA. 2019. PMID: 31110137 Free PMC article. - A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence.
Liang WW, Cheng SC. Liang WW, et al. Genes Dev. 2015 Jan 1;29(1):81-93. doi: 10.1101/gad.253708.114. Genes Dev. 2015. PMID: 25561497 Free PMC article. - Helicases involved in splicing from malaria parasite Plasmodium falciparum.
Tuteja R. Tuteja R. Parasitol Int. 2011 Dec;60(4):335-40. doi: 10.1016/j.parint.2011.09.007. Epub 2011 Oct 1. Parasitol Int. 2011. PMID: 21996352 Review. - Flipping the switch to an active spliceosome.
Murray HL, Jarrell KA. Murray HL, et al. Cell. 1999 Mar 5;96(5):599-602. doi: 10.1016/s0092-8674(00)80568-1. Cell. 1999. PMID: 10089873 Review. No abstract available.
Cited by
- A bird's-eye view of post-translational modifications in the spliceosome and their roles in spliceosome dynamics.
McKay SL, Johnson TL. McKay SL, et al. Mol Biosyst. 2010 Nov;6(11):2093-102. doi: 10.1039/c002828b. Epub 2010 Jul 29. Mol Biosyst. 2010. PMID: 20672149 Free PMC article. - Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase.
Kistler AL, Guthrie C. Kistler AL, et al. Genes Dev. 2001 Jan 1;15(1):42-9. doi: 10.1101/gad.851301. Genes Dev. 2001. PMID: 11156604 Free PMC article. - Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways.
Chen L, Weinmeister R, Kralovicova J, Eperon LP, Vorechovsky I, Hudson AJ, Eperon IC. Chen L, et al. Nucleic Acids Res. 2017 Feb 28;45(4):2051-2067. doi: 10.1093/nar/gkw860. Nucleic Acids Res. 2017. PMID: 27683217 Free PMC article. - Using yeast genetics to study splicing mechanisms.
Hossain MA, Johnson TL. Hossain MA, et al. Methods Mol Biol. 2014;1126:285-98. doi: 10.1007/978-1-62703-980-2_21. Methods Mol Biol. 2014. PMID: 24549672 Free PMC article. - Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing.
Perriman RJ, Ares M Jr. Perriman RJ, et al. Genes Dev. 2007 Apr 1;21(7):811-20. doi: 10.1101/gad.1524307. Genes Dev. 2007. PMID: 17403781 Free PMC article.
References
- Ares M, Jr, Igel AH. Lethal and temperature sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes & Dev. 1990;4:2132–2145. - PubMed
- Ares M, Jr, Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog Nucleic Acid Res. 1995;50:131–159. - PubMed
- Behrens SE, Galsson F, Legrain P, Luhrmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci. 1993;90:8229–8233. - PMC - PubMed
- Brosi R, Groning K, Behrens SE, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases