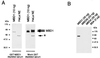Active repression of methylated genes by the chromosomal protein MBD1 - PubMed (original) (raw)
Active repression of methylated genes by the chromosomal protein MBD1
H H Ng et al. Mol Cell Biol. 2000 Feb.
Abstract
MBD1 belongs to a family of mammalian proteins that share a methyl-CpG binding domain. Previous work has shown that MBD1 binds to methylated sites in vivo and in vitro and can repress transcription from methylated templates in transcription extracts and in cultured cells. In the present study we established by several experimental criteria that, contrary to a previous report, MBD1 is not a component of the MeCP1 repressor complex. We identified a powerful transcriptional repression domain (TRD) at the C terminus of MBD1 that can actively repress transcription at a distance. Methylation-dependent repression in vivo depends on the presence of both the TRD and the methyl-CpG binding domain. The mechanism is likely to involve deacetylation, since the deacetylase inhibitor trichostatin A can overcome MBD1-mediated repression. Accordingly, we found that endogenous MBD1 is particularly concentrated at sites of centromeric heterochromatin, where acetylated histone H4 is deficient. Unlike MBD2 and MeCP2, MBD1 is not depleted by antibodies to the histone deacetylase HDAC1. Thus, the deacetylase-dependent pathway by which MBD1 actively silences methylated genes is likely to be different from that utilized by the methylation-dependent repressors MeCP1 and MeCP2.
Figures
FIG. 1
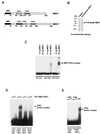
Binding to CpG methylated DNA by the methyl-CpG binding domain of MBD1. (A) Two forms of MBD1 that can be expressed by the human gene (19): MBD1 (accession no. Y10746) and MBD1* (accession no. AAD50371). Both have an N-terminal methyl-CpG binding domain (black box) and two or three CxxC domains (shaded boxes). (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of recombinant full-length MBD1 stained with Coomassie blue. (C) Bandshift assays of full-length MBD1 in the presence of differentially methylated CG11 probes. Probes were either nonmethylated (M−), or methylated at 7 _Hpa_II sites (HpaII CG11) or 20 _Hha_I sites (HhaI CG11) and labeled with 32P. (D) A 67-amino-acid region of the methyl-CpG binding domain fused to GST (N67) binds to symmetrically methylated CpGs but not to nonmethylated or hemimethylated CpGs. (E) Amino acid substitution of the conserved proline 38 residue (P38K) abolishes methyl-CpG binding. WT, wild type.
FIG. 2
Characterization of an antibody raised against the C terminus of MBD1. (A) Anti-MBD1 antibody detects two major forms of MBD1 in HeLa nuclear extract (thick arrows; right panel). Adsorption of the antibody with the cognate antigen (GST-MBD1) drastically reduces the reaction of the serum with recombinant MBD1 and prevents the detection of MBD1 in a HeLa nuclear extract (left panel). Mock adsorption with GST does not affect the reactivity of the serum toward MBD1. The asterisk denotes a nonspecific band due to secondary antibody. HeLa NE denotes 30 μg of HeLa nuclear extract. (B) Anti-MBD1 antibody recognizes MBD1 but not MBD2b, MBD4, or bovine serum albumin.
FIG. 3
MBD1 is not a component of HeLa MeCP1 activity. (A) Extracts subjected to adsorption by anti-MBD1 antibodies (immune) contained reduced levels of MBD1, whereas preimmune antibodies did not deplete MBD1. Only immunoprecipitates by anti-MBD1 antibodies contained MBD1. We used 1, 5, and 10 μl of sera for the experiments. (B) Both MBD1-immunodepleted extracts (I) and mock-depleted extracts (PI) retained MeCP1 activity, as assayed by the formation of a complex with methylated probe DNA. MeCG11 is the methylated version of CG11. (C) Antibody against MBD1 did not supershift MeCP1 activity. Serum was not added in lanes 2 and 8. Lanes 3, 5, 9, and 11 contain 1 μl of preimmune or immune serum. Half the amount of serum was used in lanes 4, 6, 10, and 12. The complexes formed with the nonmethylated probe are nonspecific complexes not due to MeCP1 (7). (D) MBD1 and MBD2 are not associated in the same complex. Anti-MBD1 antibodies did not immunoprecipitate MBD2, and anti-MBD2 antibodies did not immunoprecipitate MBD1. (E) MBD1 is not associated with the MBD2-HDAC1 MeCP1 complex. Anti-HDAC1 antibodies immunodeplete MBD2 but not MBD1. Anti-MTA2 and anti-SAP30 antibodies were used as controls, since neither MBD1 nor MBD2 associates with these proteins. (F) Apparent molecular masses of MBD1 and MBD2. HeLa nuclear extract was applied to a Superose 6 gel filtration column. Eluted fractions were analyzed by Western blotting to detect MBD1 and MBD2.
FIG. 4
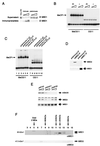
Effect of regions of MBD1 on transcription when tethered to the DNA by a GAL4 DNA binding domain. (A) Map of reporters with five GAL4 binding sites (G5) and no GAL4 binding sites (G0) and of the effectors which express a fusion between the GAL4 binding site and regions of MBD1 downstream. Effector proteins were expressed from a CMV promoter. (B) Examples of the relative transcription levels of G5 (squares) and G0 (circles) reporters in the presence of increasing amounts of different GAL4-MBD1 effectors. The MBD1(273–340) profile was obtained with effector constructs that contained the second CxxC domain, whereas the MBD1(506–538) profile was obtained with effectors containing the C terminus of MBD1. The shaded box next to each panel provides a key to the repression profile obtained with the effector constructs listed in panel C. (C) Summary maps showing regions in MBD1 that can affect the transcription of reporters in the above assay. Repression characteristic of the CxxC domain [see MBD1(273–340) above] was seen with grey constructs, whereas repression characteristic of the C terminus [see MBD1(506–538) above] was seen with diagonally shaded constructs. aa, amino acids.
FIG. 5
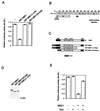
Repression by the C-terminal TRD acts at a distance but is sensitive to TRD mutations and the presence of TSA. (A) Amino acid sequence conservation of the TRD between human and mouse MBD1s. Mutations of hydrophobic residue isoleucine-527 (I527R) and leucine-530 (L530R) destroys the repression activity of the TRD, but mutation of threonine-525 (T525K) leads to partial loss of repression activity. Inset: wild-type (WT) and mutant GAL4-TRD fusion proteins are expressed at equal levels. (B) GAL4-TRD can repress transcription from a distance. Repression was effective when GAL4 binding sites were placed at 400, 1,300, and 2,100 bp from the transcription start site in the reporter construct. (C) TSA (100 ng/ml) can partially relieve repression by GAL4-TRD. The results shown are based on three independent transfections.
FIG. 6
Repression of a methylated reporter gene by MBD1 depends on the TRD and methyl-CpG binding domains and is sensitive to TSA. (A) Mouse L929 cells were transfected with an SV40-luciferase reporter (2 μg) that was nonmethylated (M−) or methylated at 25 _Hha_I (GCGC) sites (M+). Levels of M+ and M− reporter expression (luciferase activity) were normalised to the expression level of cotransfected CMV β-galactosidase control reporter (1 μg). Transcription levels were expressed as the ratio of M+ to M− normalised expression levels. The low density of methylation in the pGL2 control reporter had a negligible effect on transcription in the absence of cotransfected MBD1 but caused repression relative to the nonmethylated reporter in the presence of 0.5 μg of intact MBD1 fused to a Myc epitope tag (5MT-MBD1). N-terminal or C-terminal deletions prevented repression. (B) Diagram of the pGL2 SV40-luciferase reporter, showing the locations of methylated _Hha_I sites. (C) Diagram of the 5MT-MBD1 proteins used to obtain data shown in panel A. (D) Western blots of proteins extracted from cells transfected by each of the 5MT-MBD1 constructs showing equivalent expression. The blots were probed with anti-Myc monoclonal antibody 9e10. (E) TSA (100 ng/ml) overcomes repression by MBD1 of a methylated reporter gene. The results shown are based on three independent transfections.
FIG. 7
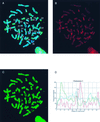
Localization of MBD1 on human metaphase chromosomes in relation to the distribution of histone H4 acetylation. (A) Unfixed cytospin HF19 primary female fibroblast metaphase cell, simultaneously immunolabelled with sheep S751 anti-serum (detected in red) and rabbit anti-acetyl(Lys-12)-H4 (detected in green), counterstained for DNA with Hoechst 33258 (blue). Numbered chromosomes were identified from their Hoechst fluorescence and H4-acetylation immunofluorescence patterns. Xa and Xi refer to the active and inactive X chromosomes, respectively. (B) Separated red image showing the MBD1 concentration at centromeres (particularly noticeable in constitutive pericentromeric heterochromatin of chromosomes 1, 9, 15 and 16 [see also panel A]) and weaker, nonuniform labelling of euchromatic chromosome arms. Specific euchromatin localization is evident, since both chromatids exhibit a similar pattern of immunofluorescent bands (this is more apparent in some chromosomes than others). (C) Separated green image showing the histone H4 acetylation profile and identifying hypoacetylated constitutive heterochromatin and the inactive X chromosome (Xi). (D) The intensities of MBD1 and acetylated H4 immunofluorescence (red and green, respectively) and Hoechst 33258 DNA fluorescence (blue) are plotted along the axis of the nonoverlapped chromosome 1 from panel A. (The position along the chromosome axis is given in arbitrary units, starting at the tip of the short arm; fluorescence intensity is likewise given in arbitrary units.) The concentration of MBD1 at the centromere is clearly indicated by the peak in the red trace, in contrast to the very low level of histone H4 acetylation in the same region (green trace). MBD1 fluorescence along the chromosome arms shows much weaker maxima.
Similar articles
- Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex.
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nan X, et al. Nature. 1998 May 28;393(6683):386-9. doi: 10.1038/30764. Nature. 1998. PMID: 9620804 - MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing.
Jin SG, Jiang CL, Rauch T, Li H, Pfeifer GP. Jin SG, et al. J Biol Chem. 2005 Apr 1;280(13):12700-9. doi: 10.1074/jbc.M413492200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15701600 - Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression.
Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Fujita N, et al. J Biol Chem. 2003 Jun 27;278(26):24132-8. doi: 10.1074/jbc.M302283200. Epub 2003 Apr 23. J Biol Chem. 2003. PMID: 12711603 - Methyl-CpG-binding proteins. Targeting specific gene repression.
Ballestar E, Wolffe AP. Ballestar E, et al. Eur J Biochem. 2001 Jan;268(1):1-6. doi: 10.1046/j.1432-1327.2001.01869.x. Eur J Biochem. 2001. PMID: 11121095 Review. - Regulation of transcription and chromatin by methyl-CpG binding protein MBD1.
Nakao M, Matsui S, Yamamoto S, Okumura K, Shirakawa M, Fujita N. Nakao M, et al. Brain Dev. 2001 Dec;23 Suppl 1:S174-6. doi: 10.1016/s0387-7604(01)00348-5. Brain Dev. 2001. PMID: 11738867 Review.
Cited by
- Light-Activatable MBD-Readers of 5-Methylcytosine Reveal Domain-Dependent Chromatin Association Kinetics In Vivo.
Lin TC, Engelhard L, Söldner B, Linser R, Summerer D. Lin TC, et al. Adv Sci (Weinh). 2024 Mar;11(11):e2307930. doi: 10.1002/advs.202307930. Epub 2024 Jan 2. Adv Sci (Weinh). 2024. PMID: 38164822 Free PMC article. - Dynamic regulation of estrogen receptor-alpha gene expression in the brain: a role for promoter methylation?
Wilson ME, Westberry JM, Prewitt AK. Wilson ME, et al. Front Neuroendocrinol. 2008 Jun;29(3):375-85. doi: 10.1016/j.yfrne.2008.03.002. Epub 2008 Mar 13. Front Neuroendocrinol. 2008. PMID: 18439661 Free PMC article. Review. - Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA.
Clouaire T, de Las Heras JI, Merusi C, Stancheva I. Clouaire T, et al. Nucleic Acids Res. 2010 Aug;38(14):4620-34. doi: 10.1093/nar/gkq228. Epub 2010 Apr 8. Nucleic Acids Res. 2010. PMID: 20378711 Free PMC article. - Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment.
Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N. Cooper S, et al. Cell Rep. 2014 Jun 12;7(5):1456-1470. doi: 10.1016/j.celrep.2014.04.012. Epub 2014 May 22. Cell Rep. 2014. PMID: 24857660 Free PMC article. - An epigenetic regulator: methyl-CpG-binding domain protein 1 (MBD1).
Li L, Chen BF, Chan WY. Li L, et al. Int J Mol Sci. 2015 Mar 5;16(3):5125-40. doi: 10.3390/ijms16035125. Int J Mol Sci. 2015. PMID: 25751725 Free PMC article. Review.
References
- Barbin A, Montpellier C, Kokalj-Vokac N, Gibaud A, Niveleau A, Malfoy B, Dutrillaux B, Bourgeois C A. New sites of methylcytosine-rich DNA detected on metaphase chromosomes. Hum Genet. 1994;94:684–692. - PubMed
- Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. - PubMed
- Bestor T H, Walsh C P, Yoder J A. Does DNA methylation control transposition of selfish elements in the germline. Trends Genet. 1997;13:470–472. - PubMed
- Bird A, Wolffe A P. Methylation-induced repression—belts, braces and chromatin. Cell. 1999;99:451–454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous