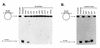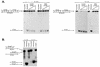Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase - PubMed (original) (raw)
Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase
Y Hirota et al. Nucleic Acids Res. 2000.
Abstract
Recently, we cloned two highly related human genes, hChlR1 ( DDX11 ) and hChlR2 ( DDX12 ), which appear to be homologs of the Saccharomyces cerevisiae CHL1 gene. Nucleotide sequence analysis suggests that these genes encode new members of the DEAH family of DNA helicases. While the enzymatic activity of CHL1 has not been characterized, the protein is required for the maintenance of high fidelity chromosome segregation in yeast. Here we report that the hChlR1 protein is a novel human DNA helicase. We have expressed and purified hChlR1 using a baculovirus system and analyzed its enzymatic activity. The recombinant hChlR1 protein possesses both ATPase and DNA helicase activities that are strictly dependent on DNA, divalent cations and ATP. These activities are abolished by a single amino acid substitution in the ATP-binding domain. The hChlR1 protein can unwind both DNA/DNA and RNA/DNA substrates. It has a preference for movement in the 5'-->3' direction on short single-stranded DNA templates. However, unlike other DNA helicases, the hChlR1 DNA helicase can translocate along single-stranded DNA in both directions when substrates have a very long single-stranded DNA region. The enzymatic activities of hChlR1 suggest that DNA helicases are required for maintaining the fidelity of chromosome segregation.
Figures
Figure 1
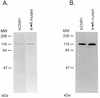
Purification of wild-type and mutant hChlR1 proteins by Ni2+ column chromatography. High Five insect cells were infected with baculoviruses carrying either the wild-type or a mutant hChlR1 cDNA with a single amino acid change in the ATP-binding site. Forty-eight hours after infection the cells were collected and lysed. Both the proteins were purified by Ni2+ affinity column chromatography. The purified proteins were electrophoresed through SDS–polyacrylamide gels. (A) The proteins were stained with Coomassie brilliant blue. The following concentrations of hChlR1 proteins were loaded on the gel: lane 1, 150 ng of hChlR1; lane 2, 300 ng of the K-R mutant. (B) Western blotting using affinity-purified Hel1 antisera which recognizes the N-terminus of the hChlR1 protein. Lane 1, 7.5 ng of hChlR1; lane 2, 15 ng of the K-R mutant.
Figure 2
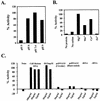
Wild-type hChlR1 protein, but not mutant hChlR1 protein, hydrolyzes ATP in the presence of calf thymus DNA. ATPase activity was detected by measuring the release of inorganic phosphate from [γ-32P]ATP. (A) Release of free phosphate during a 150 min assay. The results are expressed as the means ± SD of the experiment performed in triplicate. Diamond, no hChlR1 protein; closed circle, wild-type hChlR1 protein with 100 µg/ml calf thymus DNA; open circle, wild-type hChlR1 protein without calf thymus DNA; closed square, mutant hChlR1 protein with 100 µg/ml calf thymus DNA; open square, mutant hChlR1 protein without calf thymus DNA. (B) Comparison of the specific activities of the wild-type and mutant hChlR1 proteins. The results were calculated using means of the triplicate samples from the experiments. Closed bar, in the presence of 100 µg/ml calf thymus DNA; open bar, in the absence of calf thymus DNA.
Figure 3
Characterization of hChlR1 ATPase activity. ATPase activity was detected in reaction buffer(s) at several different pH (A), containing different cations (B) or containing different types of nucleic acids (C). These experiments were performed in duplicate. The results are shown as the percentage of the value obtained under standard experimental conditions (pH 7.4, Mg2+ and 100 µg/ml calf thymus DNA).
Figure 4
The wild-type hChlR1 protein, but not mutant hChlR1 protein, unwinds DNA/DNA duplexes. Helicase activity was detected by separating the labeled oligonucleotide displaced from a partial duplex substrate on a non-denaturing polyacrylamide gel. The structure of the substrate is shown on the left side. Asterisks denote the 32P-labeled end. The heat-denatured substrate was applied in the second lane shown as Denatured. –ATP and –Mg2+ indicate that ATP or Mg2+, respectively, was removed from the reaction (lane 6), indicating that the DNA helicase activity of hChlR1 protein requires ATP and Mg2+.
Figure 5
Substrate dependence of hChlR1 DNA helicase activity. Helicase activity was detected using different types of substrates. The structure of the substrate is shown at the left side of each panel. Asterisks denote the 32P-labeled end. The heat-denatured substrate was applied in lanes marked Denatured. (A) Three oligonucleotide substrates of different lengths were annealed to the M13mp18 DNA and used in the helicase assay. (B) RNA/DNA heteroduplexes, which consisted of an 18mer RNA oligonucleotide annealed to M13mp18 DNA, and DNA/DNA duplexes were used as helicase substrates. The sequence of the 18mer RNA oligonucleotide was identical to that of the 18mer DNA. (C) A 19mer blunt-ended DNA substrate prepared by fill-in reaction with Klenow fragment (exo–).
Figure 6
Nucleotide and cation dependence of hChlR1 DNA helicase activity. (A) All of the normal nucleoside 5′-triphosphates and an unhydrolyzable ATP analog, ATPγS, were used as cofactors of the hChlR1 DNA helicase. The concentration of nucleotide added to the reaction buffer was 1 mM. (B) Reaction buffers including different divalent cations at a concentration of 1 mM were used for helicase assay. The structure of the substrate is shown at the left side of each panel. Asterisks denote the 32P-labeled end. The heat-denatured substrate was applied in lanes marked Denatured.
Figure 7
Directionality of hChlR1 DNA helicase. The linearized M13mp18 DNAs with blunt-ended partially duplexed termini were used as substrates (A). To distinguish either 3′→5′ or 5′→3′ directional movement, the oligonucleotides annealed to M13mp18 DNA were labeled as shown on the left side of each panel. WRN helicase, which is known to translocate only in a 3′→5′ direction, was used in order to confirm that the substrates were properly prepared. (B) Substrates with a short single-stranded DNA were used to examine the directionality of hChlR1 DNA helicase. The structures of two substrates used in this experiment are shown together on the left side of the panel. Asterisks denote the 32P-labeled end. The heat-denatured substrate was applied in lanes marked Denatured.
Similar articles
- Characterization of putative human homologues of the yeast chromosome transmission fidelity gene, CHL1.
Amann J, Kidd VJ, Lahti JM. Amann J, et al. J Biol Chem. 1997 Feb 7;272(6):3823-32. doi: 10.1074/jbc.272.6.3823. J Biol Chem. 1997. PMID: 9013641 - Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1.
Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, Hurwitz J. Farina A, et al. J Biol Chem. 2008 Jul 25;283(30):20925-36. doi: 10.1074/jbc.M802696200. Epub 2008 May 21. J Biol Chem. 2008. PMID: 18499658 Free PMC article. - Characterization of Plasmodium falciparum ATP-dependent DNA helicase RuvB3.
Limudomporn P, Moonsom S, Leartsakulpanich U, Suntornthiticharoen P, Petmitr S, Weinfeld M, Chavalitshewinkoon-Petmitr P. Limudomporn P, et al. Malar J. 2016 Nov 3;15(1):526. doi: 10.1186/s12936-016-1573-2. Malar J. 2016. PMID: 27809838 Free PMC article. - Rapid purification of helicase proteins and in vitro analysis of helicase activity.
Tahmaseb K, Matson SW. Tahmaseb K, et al. Methods. 2010 Jul;51(3):322-8. doi: 10.1016/j.ymeth.2010.02.008. Epub 2010 Feb 12. Methods. 2010. PMID: 20153831 Free PMC article. Review. - The Genome Stability Maintenance DNA Helicase DDX11 and Its Role in Cancer.
Mahtab M, Boavida A, Santos D, Pisani FM. Mahtab M, et al. Genes (Basel). 2021 Mar 10;12(3):395. doi: 10.3390/genes12030395. Genes (Basel). 2021. PMID: 33802088 Free PMC article. Review.
Cited by
- A distinct triplex DNA unwinding activity of ChlR1 helicase.
Guo M, Hundseth K, Ding H, Vidhyasagar V, Inoue A, Nguyen CH, Zain R, Lee JS, Wu Y. Guo M, et al. J Biol Chem. 2015 Feb 20;290(8):5174-5189. doi: 10.1074/jbc.M114.634923. Epub 2015 Jan 5. J Biol Chem. 2015. PMID: 25561740 Free PMC article. - DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster.
Wu Y, Brosh RM Jr. Wu Y, et al. Nucleic Acids Res. 2012 May;40(10):4247-60. doi: 10.1093/nar/gks039. Epub 2012 Jan 28. Nucleic Acids Res. 2012. PMID: 22287629 Free PMC article. Review. - Maintenance of genome integrity by the late-acting cytoplasmic iron-sulfur assembly (CIA) complex.
Petronek MS, Allen BG. Petronek MS, et al. Front Genet. 2023 Mar 8;14:1152398. doi: 10.3389/fgene.2023.1152398. eCollection 2023. Front Genet. 2023. PMID: 36968611 Free PMC article. Review. - Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome.
Bharti SK, Khan I, Banerjee T, Sommers JA, Wu Y, Brosh RM Jr. Bharti SK, et al. Cell Mol Life Sci. 2014 Jul;71(14):2625-39. doi: 10.1007/s00018-014-1569-4. Epub 2014 Feb 1. Cell Mol Life Sci. 2014. PMID: 24487782 Free PMC article. Review. - Put on your thinking cap: G-quadruplexes, helicases, and telomeres.
Brosh RM Jr. Brosh RM Jr. Aging (Albany NY). 2011 Apr;3(4):332-5. doi: 10.18632/aging.100307. Aging (Albany NY). 2011. PMID: 21732565 Free PMC article. No abstract available.
References
- Lohman T.M. and Bjornson,K.P. (1996) Annu. Rev. Biochem., 65, 169–214. - PubMed
- Matson S.W. and Kaiser-Rogers,K.A. (1990) Annu. Rev. Biochem., 59, 289–329. - PubMed
- Borowiec J.A. (1996) In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 545–574.
- Ellis N.A. (1997) Curr. Opin. Genet. Dev., 7, 354–363. - PubMed
- Hodgman T.C. (1988) Nature, 333, 22–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous