Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity - PubMed (original) (raw)
Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity
F von Wintzingerode et al. Appl Environ Microbiol. 2000 Feb.
Abstract
Peptide nucleic acid (PNA)-mediated PCR clamping (H. Orum, P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt, and C. Stanley, Nucleic Acids Res. 21:5332-5336, 1993) was introduced as a novel procedure to selectively amplify ribosomal DNAs (rDNAs) which are not frequently found in clone libraries generated by standard PCR from complex microbial consortia. Three different PNA molecules were used; two of these molecules (PNA-ALF and PNA-EUB353) overlapped with one of the amplification primers, whereas PNA-1114F hybridized to the middle of the amplified region. Thus, PCR clamping was achieved either by competitive binding between the PNA molecules and the forward or reverse primers (competitive clamping) or by hindering polymerase readthrough (elongation arrest). Gene libraries generated from mixed rDNA templates by using PCR clamping are enriched for clones that do not contain sequences homologous to the appropriate PNA oligomer. This effect of PCR clamping was exploited in the following two ways: (i) analysis of gene libraries generated by PCR clamping with PNA-ALF together with standard libraries reduced the number of clones which had to be analyzed to detect all of the different sequences present in an artificial rDNA mixture; and (ii) PCR clamping with PNA-EUB353 and PNA-1114F was used to selectively recover rDNA sequences which represented recently described phylogenetic groups (NKB19, TM6, cluster related to green nonsulfur bacteria) from an anaerobic, dechlorinating consortium described previously. We concluded that PCR clamping might be a useful supplement to standard PCR amplification in rDNA-based studies of microbial diversity and could be used to selectively recover members of undescribed phylogenetic clusters from complex microbial communities.
Figures
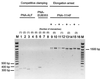
FIG. 1
Agarose gel electrophoresis analysis of products of various PCR clamping reactions. Lanes 1 through 5, PCR performed with PNA-ALF and reference rDNAs (V. spinosum [one mismatch], SJA-186 [two mismatches], SJA-53 [one mismatch], SJA-9 [no mismatch], SJA-105 [no mismatch]); lanes 6 through 8, PCR performed with PNA-EUB353 and reference rDNAs (V. spinosum [two mismatches], strain OLB-1 [no mismatch], C. gallinarum [no mismatch]); lanes 9 through 16, duplicate PCR performed with (+) or without (−) PNA-1114F and reference rDNAs (V. spinosum [no mismatch], strain OLB-1 [one mismatch], SJA-131 [two mismatches], SJA-4 [three mismatches]); lanes M, length standard (100 bp plus; MBI-Fermentas, St.-Leon Roth, Germany). The sizes of relevant bands are shown on the left and right.
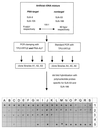
FIG. 2
Analysis of an artificial rDNA mixture by competitive clamping: schematic diagram of PCR clamping performed with PNA-ALF and results of dot blot hybridizations with polynucleotide probes specific for SJA-53 and SJA-186. Positions P1 to K3, selected clones from libraries A1 to A3 (PCR clamping); positions L3 to G5, selected clones from libraries A4 to A6 (control PCR). The following 16S rDNAs were used as controls: SJA-9 (position I5), SJA-53 (position J5), SJA-105 (position K5), and SJA-186 (position M5). Asterisks indicate positions where no samples were applied.
FIG. 3
Schematic diagram of PNA-mediated PCR clamping of 16S rDNA amplification. (A) Competitive clamping: inhibition of PCR amplification by PNA-ALF-mediated exclusion of primer TPU1. (B) Elongation arrest: inhibition of PCR amplification by binding of PNA-1114F to an internal target sequence, which prevents readthrough by the Taq polymerase. In both cases amplification proceeds only if one or more base substitutions in the binding sites of appropriate PNA molecules are present.
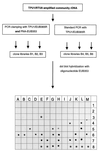
FIG. 4
Analysis of community rDNA by competitive clamping: schematic diagram of PCR clamping performed with PNA-EUB353 and results of dot blot hybridizations with oligonucleotide probe EUB353. Positions A1 to H4, selected clones from libraries B1 to B3 (PCR clamping); positions I4 to A8, selected clones from libraries B4 to B6 (standard PCR). The following 16S rDNAs were used as controls: SJA-22 (position B8), SJA-43 (position C8), SJA-131 (position D8), Pseudomonas aeruginosa ATCC 25330 (position G8), C. gallinarum DSM 4847T (position H8), and V. spinosum DSM 4136T (position I8). Asterisks indicate positions where no samples were applied.
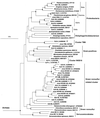
FIG. 5
Phylogenetic dendrogram showing clusters mentioned in this study. For sequences marked with an asterisk a corresponding rDNA clone (level of sequence similarity, >99%) was recovered by competitive clamping with PNA-EUB353 (Table 5). For sequences marked with a less-than sign the corresponding rDNA clone was obtained by elongation arrest with PNA-1114F. PNA-1114F nontarget clones C1-28 and C2-12 could not be affiliated with any of the previously described SJA clone families in the cluster related to GNS bacteria and were included in the dendrogram. 16S rDNA clone sequences SJP-2 and SJP-3 (∼1,300 bp) were amplified from total community rDNA by a seminested PCR performed with forward primer NKB19F (5′ GCTGCAAGGCGTCGCCG 3′) derived from NKB19-related clones B1-9, B1-11, and B3-6 and universal reverse primer RTU8. Branch points supported by bootstrap values of >74% are indicated by solid circles. Open circles indicate branch points supported by bootstrap values between 50 and 74%. Branch points without circles were not resolved (bootstrap values, <50%). Scale bar = 10% sequence divergence.
Similar articles
- Application of peptide nucleic acid (PNA)-PCR clamping technique to investigate the community structures of rhizobacteria associated with plant roots.
Sakai M, Ikenaga M. Sakai M, et al. J Microbiol Methods. 2013 Mar;92(3):281-8. doi: 10.1016/j.mimet.2012.09.036. Epub 2013 Jan 10. J Microbiol Methods. 2013. PMID: 23313555 - Peptide Nucleic Acid (PNA) Clamps to Reduce Co-amplification of Plant DNA During PCR Amplification of 16S rRNA Genes from Endophytic Bacteria.
Kawasaki A, Ryan PR. Kawasaki A, et al. Methods Mol Biol. 2021;2232:123-134. doi: 10.1007/978-1-0716-1040-4_11. Methods Mol Biol. 2021. PMID: 33161544 - Chloroplast sequence variation and the efficacy of peptide nucleic acids for blocking host amplification in plant microbiome studies.
Fitzpatrick CR, Lu-Irving P, Copeland J, Guttman DS, Wang PW, Baltrus DA, Dlugosch KM, Johnson MTJ. Fitzpatrick CR, et al. Microbiome. 2018 Aug 18;6(1):144. doi: 10.1186/s40168-018-0534-0. Microbiome. 2018. PMID: 30121081 Free PMC article. - A survey of the methods for the characterization of microbial consortia and communities.
Spiegelman D, Whissell G, Greer CW. Spiegelman D, et al. Can J Microbiol. 2005 May;51(5):355-86. doi: 10.1139/w05-003. Can J Microbiol. 2005. PMID: 16088332 Review. - Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis.
von Wintzingerode F, Göbel UB, Stackebrandt E. von Wintzingerode F, et al. FEMS Microbiol Rev. 1997 Nov;21(3):213-29. doi: 10.1111/j.1574-6976.1997.tb00351.x. FEMS Microbiol Rev. 1997. PMID: 9451814 Review.
Cited by
- Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing.
Song L, Xie K. Song L, et al. Microbiome. 2020 Jun 3;8(1):80. doi: 10.1186/s40168-020-00859-0. Microbiome. 2020. PMID: 32493511 Free PMC article. - Characterizing Microbiomes via Sequencing of Marker Loci: Techniques To Improve Throughput, Account for Cross-Contamination, and Reduce Cost.
Harrison JG, Randolph GD, Buerkle CA. Harrison JG, et al. mSystems. 2021 Aug 31;6(4):e0029421. doi: 10.1128/mSystems.00294-21. Epub 2021 Jul 13. mSystems. 2021. PMID: 34254828 Free PMC article. - Depletion of unwanted nucleic acid templates by selective cleavage: LNAzymes, catalytically active oligonucleotides containing locked nucleic acids, open a new window for detecting rare microbial community members.
Dolinsek J, Dorninger C, Lagkouvardos I, Wagner M, Daims H. Dolinsek J, et al. Appl Environ Microbiol. 2013 Mar;79(5):1534-44. doi: 10.1128/AEM.03392-12. Epub 2012 Dec 21. Appl Environ Microbiol. 2013. PMID: 23263968 Free PMC article. - Comparison of RNA- and DNA-based 16S amplicon sequencing to find the optimal approach for the analysis of the uterine microbiome.
Dyroff AI, López-Valiñas Á, Magalhaes HB, Podico G, Canisso IF, Almiñana C, Bauersachs S. Dyroff AI, et al. Sci Rep. 2025 May 16;15(1):17037. doi: 10.1038/s41598-025-00969-5. Sci Rep. 2025. PMID: 40379732 Free PMC article. - Suicide polymerase endonuclease restriction, a novel technique for enhancing PCR amplification of minor DNA templates.
Green SJ, Minz D. Green SJ, et al. Appl Environ Microbiol. 2005 Aug;71(8):4721-7. doi: 10.1128/AEM.71.8.4721-4727.2005. Appl Environ Microbiol. 2005. PMID: 16085868 Free PMC article.
References
- Behn M, Schuermann M. Simple and reliable factor V genotyping by PNA-mediated PCR-clamping. Thromb Haemostasis. 1998;79:773–777. - PubMed
- Boyle J S, Lew A M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995;11:8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases