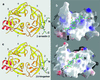Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin - PubMed (original) (raw)
Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin
E ter Haar et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The "WD40" domain is a widespread recognition module for linking partner proteins in intracellular networks of signaling and sorting. The clathrin amino-terminal domain, which directs incorporation of cargo into coated pits, is a beta-propeller closely related in structure to WD40 modules. The crystallographically determined structures of complexes of the clathrin-terminal domain with peptides derived from two different cargo adaptors, beta-arrestin 2 and the beta-subunit of the AP-3 complex, reveal strikingly similar peptide-in-groove interactions. The two peptides in our structures contain related, five-residue motifs, which form the core of their contact with clathrin. A number of other proteins involved in endocytosis have similar "clathrin-box" motifs, and it therefore is likely that they all bind the terminal domain in the same way. We propose that a peptide-in-groove interaction is an important general mode by which beta-propellers recognize specific target proteins.
Figures
Figure 1
Structures of clathrin-terminal domain complexed with clathrin-box peptides from β-arrestin 2 (a and b) and β3-hinge of AP-3 (c and d). (a and c) Ribbon diagrams of the complexes. The representation is a “top” view of the td40 fragment (residues 3–359) looking from the membrane toward the surrounding clathrin coat. The propeller blades are numbered 1–7; each blade contains four antiparallel β-strands, labeled a–d. Ordered portions of the bound peptides from β-arrestin 2 and β3-hinge (of sequence DTNLIEFE and VSLLDLD) are in green. The figure was made with
ribbons
(31). (b and d) Surface representation of the complexes. The images of b and d have been rotated approximately 90° toward the viewer. The map shows positive (blue), negative (red), and hydrophobic/neutral patches (white) projected onto the surface representation. The figure was made with
grasp
(32).
Figure 2
Alignment of sequences found in proteins that bind clathrin (, , –35, 38, 39). The comparison delineates a conserved clathrin-box motif, a consensus noted in ref. . The clathrin box is surrounded in each of these proteins by unrelated residues.
Figure 3
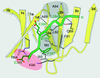
Close-up view of the peptide-in-groove interactions between the clathrin box in the peptide of β-arrestin 2 and the clathrin-terminal domain. The residues that contribute to the interaction between the β-arrestin 2 peptide and the clathrin groove between blades 1 and 2 are labeled by single-letter code and by the positions in their respective sequences. The dashed lines show hydrogen bonds between the backbones of blade 1d and bound peptide and between peptide backbone and the side chain of clathrin Q89. The locations for the two hydrophobic (gray) and polar (rose) pockets in the clathrin groove are approximate.
Figure 4
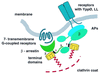
Schematic representation of the network of contact arrays of target proteins and clathrin within a coat. Key features illustrated are: 1, the contact between the clathrin terminal domain (yellow) and the clathrin-box sequences (black) in β-arrestins (green) and the β-chain of AP complexes (blue); 2, a possible additional contact (see text) between β-arrestins and terminal domain; 3, a contact, inferred from mutational studies between the carboxyl-terminal part of β-arrestins and the β-subunit of AP-2. APs are generalized adaptors for sorting signals (YppØ, LL) in various receptors; β-arrestins are specific adaptors for seven-transmembrane, G protein-coupled receptors (dark blue). The membrane is shown in gray; the framework of the clathrin coat is shown in red.
Comment in
- A groovy new structure.
Neer EJ, Smith TF. Neer EJ, et al. Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):960-2. doi: 10.1073/pnas.97.3.960. Proc Natl Acad Sci U S A. 2000. PMID: 10655467 Free PMC article. No abstract available.
Similar articles
- Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker.
ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. ter Haar E, et al. Cell. 1998 Nov 13;95(4):563-73. doi: 10.1016/s0092-8674(00)81623-2. Cell. 1998. PMID: 9827808 Free PMC article. - Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly.
Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Edeling MA, et al. Dev Cell. 2006 Mar;10(3):329-42. doi: 10.1016/j.devcel.2006.01.016. Dev Cell. 2006. PMID: 16516836 - A groovy new structure.
Neer EJ, Smith TF. Neer EJ, et al. Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):960-2. doi: 10.1073/pnas.97.3.960. Proc Natl Acad Sci U S A. 2000. PMID: 10655467 Free PMC article. No abstract available. - Endocytosis: an assembly protein for clathrin cages.
McMahon HT. McMahon HT. Curr Biol. 1999 May 6;9(9):R332-5. doi: 10.1016/s0960-9822(99)80206-1. Curr Biol. 1999. PMID: 10330371 Review. - β-arrestins and G protein-coupled receptor trafficking.
Tian X, Kang DS, Benovic JL. Tian X, et al. Handb Exp Pharmacol. 2014;219:173-86. doi: 10.1007/978-3-642-41199-1_9. Handb Exp Pharmacol. 2014. PMID: 24292830 Free PMC article. Review.
Cited by
- Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2.
Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Morgunova E, et al. Proc Natl Acad Sci U S A. 2002 May 28;99(11):7414-9. doi: 10.1073/pnas.102185399. Proc Natl Acad Sci U S A. 2002. PMID: 12032297 Free PMC article. - Involvement of a novel ADP-ribosylation factor GTPase-activating protein, SMAP, in membrane trafficking: implications in cancer cell biology.
Tanabe K, Kon S, Natsume W, Torii T, Watanabe T, Satake M. Tanabe K, et al. Cancer Sci. 2006 Sep;97(9):801-6. doi: 10.1111/j.1349-7006.2006.00251.x. Epub 2006 Jun 29. Cancer Sci. 2006. PMID: 16805823 Free PMC article. Review. - Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly.
Herrmannová A, Daujotyte D, Yang JC, Cuchalová L, Gorrec F, Wagner S, Dányi I, Lukavsky PJ, Valásek LS. Herrmannová A, et al. Nucleic Acids Res. 2012 Mar;40(5):2294-311. doi: 10.1093/nar/gkr765. Epub 2011 Nov 15. Nucleic Acids Res. 2012. PMID: 22090426 Free PMC article. - Structure and function of WD40 domain proteins.
Xu C, Min J. Xu C, et al. Protein Cell. 2011 Mar;2(3):202-14. doi: 10.1007/s13238-011-1018-1. Epub 2011 Apr 6. Protein Cell. 2011. PMID: 21468892 Free PMC article. Review. - Knockout of the gamma subunit of the AP-1 adaptor complex in the human parasite Trypanosoma cruzi impairs infectivity and differentiation and prevents the maturation and targeting of the major protease cruzipain.
Moreira CMDN, Batista CM, Fernandes JC, Kessler RL, Soares MJ, Fragoso SP. Moreira CMDN, et al. PLoS One. 2017 Jul 31;12(7):e0179615. doi: 10.1371/journal.pone.0179615. eCollection 2017. PLoS One. 2017. PMID: 28759609 Free PMC article.
References
- Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. Cell. 1995;83:1047–1058. - PubMed
- Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. - PubMed
- Smith T F, Gaitatzes C, Saxena K, Neer E J. Trends Biochem Sci. 1999;24:181–185. - PubMed
- Ungewickell E, Branton D. Nature (London) 1981;289:420–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases