Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins - PubMed (original) (raw)
Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins
T Ito et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Protein-protein interactions play pivotal roles in various aspects of the structural and functional organization of the cell, and their complete description is indispensable to thorough understanding of the cell. As an approach toward this goal, here we report a comprehensive system to examine two-hybrid interactions in all of the possible combinations between proteins of Saccharomyces cerevisiae. We cloned all of the yeast ORFs individually as a DNA-binding domain fusion ("bait") in a MATa strain and as an activation domain fusion ("prey") in a MATalpha strain, and subsequently divided them into pools, each containing 96 clones. These bait and prey clone pools were systematically mated with each other, and the transformants were subjected to strict selection for the activation of three reporter genes followed by sequence tagging. Our initial examination of approximately 4 x 10(6) different combinations, constituting approximately 10% of the total to be tested, has revealed 183 independent two-hybrid interactions, more than half of which are entirely novel. Notably, the obtained binary data allow us to extract more complex interaction networks, including the one that may explain a currently unsolved mechanism for the connection between distinct steps of vesicular transport. The approach described here thus will provide many leads for integration of various cellular functions and serve as a major driving force in the completion of the protein-protein interaction map.
Figures
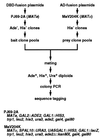
Figure 1
Outline of the strategy for comprehensive two-hybrid screening for mutually interacting proteins. The genotypes of the two strains used in the system are also shown. See text for details.
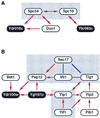
Figure 2
Complex two-hybrid interaction networks. Two-hybrid interaction networks for proteins related to spindle pole body (A) and vesicular transport (B) are shown. Red arrows indicate two-hybrid interactions, beginning from the bait and ending at the prey. Double-headed arrows mean that the interactions were detected bidirectionally. Note that arrows indicate the direction of two-hybrid interactions but not any biological orientation. Solid blue lines indicate known interactions recorded in the Yeast Proteome Database (14) but not yet detected by our two-hybrid screening. Shaded boxes are complexes or networks that were described in previous studies. Proteins of unknown function are indicated as black circles with white letters.
Similar articles
- Yeast two-hybrid screening.
Maple J, Møller SG. Maple J, et al. Methods Mol Biol. 2007;362:207-23. doi: 10.1007/978-1-59745-257-1_15. Methods Mol Biol. 2007. PMID: 17417012 - A comprehensive two-hybrid analysis to explore the yeast protein interactome.
Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Ito T, et al. Proc Natl Acad Sci U S A. 2001 Apr 10;98(8):4569-74. doi: 10.1073/pnas.061034498. Epub 2001 Mar 13. Proc Natl Acad Sci U S A. 2001. PMID: 11283351 Free PMC article. - High-throughput yeast two-hybrid assays for large-scale protein interaction mapping.
Walhout AJ, Vidal M. Walhout AJ, et al. Methods. 2001 Jul;24(3):297-306. doi: 10.1006/meth.2001.1190. Methods. 2001. PMID: 11403578 - Analysing protein-protein interactions with the yeast two-hybrid system.
Causier B, Davies B. Causier B, et al. Plant Mol Biol. 2002 Dec;50(6):855-70. doi: 10.1023/a:1021214007897. Plant Mol Biol. 2002. PMID: 12516858 Review. - Results and prospects of the yeast three-hybrid system.
Jaeger S, Eriani G, Martin F. Jaeger S, et al. FEBS Lett. 2004 Jan 2;556(1-3):7-12. doi: 10.1016/s0014-5793(03)01434-0. FEBS Lett. 2004. PMID: 14706817 Review.
Cited by
- Beyond hairballs: The use of quantitative mass spectrometry data to understand protein-protein interactions.
Gingras AC, Raught B. Gingras AC, et al. FEBS Lett. 2012 Aug 14;586(17):2723-31. doi: 10.1016/j.febslet.2012.03.065. Epub 2012 Apr 10. FEBS Lett. 2012. PMID: 22710165 Free PMC article. Review. - Biological function through network topology: a survey of the human diseasome.
Janjić V, Pržulj N. Janjić V, et al. Brief Funct Genomics. 2012 Nov;11(6):522-32. doi: 10.1093/bfgp/els037. Epub 2012 Sep 8. Brief Funct Genomics. 2012. PMID: 22962330 Free PMC article. - Yeast nuclear RNA processing.
Bernstein J, Toth EA. Bernstein J, et al. World J Biol Chem. 2012 Jan 26;3(1):7-26. doi: 10.4331/wjbc.v3.i1.7. World J Biol Chem. 2012. PMID: 22312453 Free PMC article. - Sinorhizobium meliloti YrbA binds divalent metal cations using two conserved histidines.
Roret T, Alloing G, Girardet JM, Perrot T, Dhalleine T, Couturier J, Frendo P, Didierjean C, Rouhier N. Roret T, et al. Biosci Rep. 2020 Oct 30;40(10):BSR20202956. doi: 10.1042/BSR20202956. Biosci Rep. 2020. PMID: 32970113 Free PMC article. - Genomic analysis of the hierarchical structure of regulatory networks.
Yu H, Gerstein M. Yu H, et al. Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14724-31. doi: 10.1073/pnas.0508637103. Epub 2006 Sep 26. Proc Natl Acad Sci U S A. 2006. PMID: 17003135 Free PMC article.
References
- Pawson A J, editor. Protein Modules in Signal Transduction. Berlin: Springer; 1998.
- Bartel P L, Roecklein J A, SenGupta D, Fields S. Nat Genet. 1996;12:72–77. - PubMed
- Fromont-Racine M, Rain J C, Legrain P. Nat Genet. 1997;16:277–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials