Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways - PubMed (original) (raw)
Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways
T Noda et al. J Cell Biol. 2000.
Abstract
In nutrient-rich, vegetative conditions, the yeast Saccharomyces cerevisiae transports a resident protease, aminopeptidase I (API), to the vacuole by the cytoplasm to vacuole targeting (Cvt) pathway, thus contributing to the degradative capacity of this organelle. When cells subsequently encounter starvation conditions, the machinery that recruited precursor API (prAPI) also sequesters bulk cytosol for delivery, breakdown, and recycling in the vacuole by the autophagy pathway. Each of these overlapping alternative transport pathways is specifically mobilized depending on environmental cues. The basic mechanism of cargo packaging and delivery involves the formation of a double-membrane transport vesicle around prAPI and/or bulk cytosol. Upon completion, these Cvt and autophagic vesicles are targeted to the vacuole to allow delivery of their lumenal contents. Key questions remain regarding the origin and formation of the transport vesicle. In this study, we have cloned the APG9/CVT7 gene and characterized the gene product. Apg9p/Cvt7p is the first characterized integral membrane protein required for Cvt and autophagy transport. Biochemical and morphological analyses indicate that Apg9p/Cvt7p is localized to large perivacuolar punctate structures, but does not colocalize with typical endomembrane marker proteins. Finally, we have isolated a temperature conditional allele of APG9/CVT7 and demonstrate the direct role of Apg9p/Cvt7p in the formation of the Cvt and autophagic vesicles. From these results, we propose that Apg9p/Cvt7p may serve as a marker for a specialized compartment essential for these vesicle-mediated alternative targeting pathways.
Figures
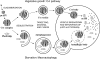
Figure 1
Model of Cvt and autophagy transport to the vacuole. There are many shared mechanistic features for the two pathways. 1, Cvt complex formation: prAPI is synthesized in the cytosol, oligomerizes into a dodecamer, and assembles into a Cvt complex; 2, double-membrane vesicle formation: the Cvt complex is enwrapped by a membrane source which, upon expansion and completion, forms the double-membraned Cvt vesicle (Cvt pathway) or autophagosome (autophagy); 3, vesicle targeting to the vacuole: the double-membrane vesicle is then targeted to the vacuole followed by fusion with the vacuolar membrane and release of the inner vesicle, termed the Cvt body (Cvt pathway) or the autophagic body (autophagy), into the vacuolar lumen. Subsequent breakdown of the Cvt or autophagic body releases prAPI into the lumen where it is proteolytically processed, in a proteinase A-dependent manner, to mature API. Major differences between the pathways include the following: 1, cargo selectivity: the Cvt pathway is a selective process and the Cvt vesicles appear to exclude bulk cytosolic components as cargo. In contrast, autophagy is nonselective and autophagosomes carry bulk cytosol, in addition to the Cvt complex, to the vacuole. prAPI is transported via both vesicle types. A recombinant, cytosolic marker protein, Pho8Δ60p, is transported to the vacuole via autophagy. Delivery of Pho8Δ60p to the vacuole can be monitored through cleavage of the COOH-terminal propeptide. 2, Vesicle size: Cvt vesicles are ∼150 nm in diameter, whereas autophagosomes are 300–900 nm in diameter. 3, Conditions for induction: the Cvt pathway is active in vegetative growth conditions while autophagy is induced under starvation conditions.
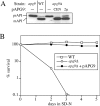
Figure 2
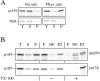
Cloning and characterization of APG9. A, Wild-type (WT; SEY6210), apg9 (THY154), _apg9_Δ (JKY007), and the _apg9_Δ strain transformed with single copy (CEN; pAPG9(414)) or multicopy (2μ; pAPG9(424)) plasmids encoding APG9 were grown to log phase in SMD. Protein extracts were prepared and analyzed by immunoblot using antiserum to API as described in Materials and Methods. The positions of precursor and mature API are indicated. The APG9 gene complements the prAPI accumulation phenotype of the _apg9_Δ mutant. B, WT (SEY6210), _apg9_Δ (JKY007), and the _apg9_Δ strain transformed with pAPG9(414) centromeric plasmid were grown in SMD and transferred to SD(−N) as described in Materials and Methods. Aliquots were removed at the indicated times and spread onto YPD plates in triplicate. Numbers of viable colonies were determined after two to three days. The APG9 gene complements the starvation-sensitive phenotype of the _apg9_Δ mutant.
Figure 3
Apg9p is an unglycosylated, integral membrane protein. A, Wild-type (WT; SEY6210), _apg9_Δ (JKY007), and the _apg9_Δ strain transformed with single copy (CEN) or multicopy (2μ) plasmids encoding APG9 were grown to log phase in SMD. Protein extracts were prepared and analyzed by immunoblot using antiserum to Apg9p. Apg9p is detected as an ∼125-kD protein. B, Hydrophobicity analysis of Apg9p based on the Kyte-Doolittle algorithm (Kyte and Doolittle 1982) predicts six to eight transmembrane domains. C, Cells of wild-type strain YW5-1B were grown in YPD, converted to spheroplasts, and osmotically lysed as described in Materials and Methods. After a preclearing step to remove cell debris, the lysates were incubated with or without 1% Triton X-100 on ice for 30 min, as indicated. Centrifugation at 10,000 g for 1 h separated the lysate into supernatant (S) and pellet (P) fractions, which were analyzed by immunoblots with antiserum to Apg9p. Apg9p is found in the low-speed pellet in the absence of Triton X-100. Detergent extraction completely solubilizes Apg9p and causes it to appear in the supernatant fraction. D, Strain CTD1 harboring a multicopy 3×HA APG9 plasmid was grown in YPD and converted to spheroplasts as described in Materials and Methods. A crude lysate was prepared and treated with or without endo H as indicated overnight at 37°C and analyzed by immunoblots with anti-HA antibodies or anti-CPY serum. Endo H treatment removes the N-linked glycosyl residues of CPY, causing a shift in its electrophoretic mobility. In contrast, Apg9p is not glycosylated and remains unchanged in the presence or absence of endo H treatment.
Figure 4
Apg9p does not comigrate with typical endomembrane markers in sucrose density gradients. A and B, Strain STY1 (_pep4_Δ) was grown in YPD to log-phase, and osmotically lysed. Lysates were precleared, loaded on a sucrose density gradient ranging from 18–55% (wt/wt), and centrifuged for 2.5 h at 174,000 g as described in Materials and Methods. Fractions were collected from the top (fraction number 1) to the bottom (fraction number 16). Fractions were subjected to SDS-PAGE and immunoblot with antiserum against Apg9p and: in A, Kex2p (trans Golgi network), Pho8p (vacuole), and Pma1p (plasma membrane); and in B, Sec12p (ER) and Pep12p (endosome). The graphs in panels A and B are from the same gradient, but are presented separately for clarity.
Figure 6
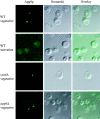
Apg9p localizes to large perivacuolar punctate structures. Immunofluorescence microscopy of strains CTD1 (_apg9_Δ), KSL12C (_vps4_Δ), and SKD6-1D (_apg6_Δ) transformed with the multicopy plasmid 3×HA APG9. Cells were grown in YPD to log phase (vegetative), and incubated further in SD(−N) medium with 1 mM PMSF for 3 h (starvation). The cells were fixed with formaldehyde and examined by immunofluorescence microscopy as described in Materials and Methods. Anti-HA and FITC-conjugated antibodies mark Apg9p (left), and the vacuole can be visualized by Nomarski optics (middle). An overlay is shown in the right panels.
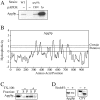
Figure 5
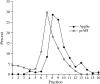
Apg9p does not comigrate with prAPI in a strain that accumulates Cvt/autophagic vesicles. Strain TK415 (_ypt7_Δ) was grown in YPD to log-phase, and osmotically lysed. Lysates were precleared, centrifuged at 100,000 g for 1 h, and the pellet fraction resuspended and centrifuged a second time. The resulting pellet fraction was subsequently loaded on a sucrose density gradient ranging from 18–55% (wt/wt), and centrifuged for 16 h at 174,000 g as described in Materials and Methods. 16 fractions were collected starting from the top and subjected to SDS-PAGE and immunoblot with antisera against Apg9p and API. The graph represents an average of two experiments.
Figure 7
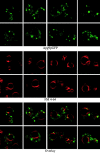
In vivo examination of the Apg9pGFP fusion protein. Wild-type (SEY6210) cells expressing Apg9pGFP from a multicopy plasmid were grown in SMD to midlog phase and labeled with FM 4-64 to stain the vacuoles as described in Materials and Methods. The labeled cells were viewed directly with a Leica DM IRB confocal microscope. Large, punctate Apg9pGFP structures appear juxtaposed to the FM 4-64–labeled vacuole membranes. In addition, smaller punctate structures appear throughout the cytoplasm.
Figure 8
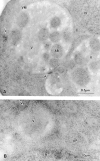
Ultrastructural localization of Apg9p for immunoelectron microscopy. Strain STY1 (_pep4_Δ) expressing 3×HA Apg9p was grown in YPD to log phase and transferred to SD(−N) medium for 2 h. The cells were fixed and stained with anti-HA antibody and 5-nm colloidal gold-conjugated goat anti–mouse IgG as described in Materials and Methods. A, Section showing autophagic bodies in the vacuole lumen. B, Section showing a cytosolic autophagosome. Apg9p is seen in patches near the vacuole, but not on autophagic bodies or autophagosomes. AB, autophagic body; AP, autophagosome; N, nucleus; V, vacuole; VM, vacuolar membrane. Arrows point to areas of concentrated Apg9p.
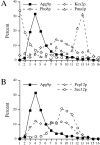
Figure 9
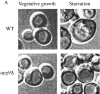
Vesicle accumulation test and kinetic analysis of prAPI import in the temperature conditional Apg9p mutant. A, Morphological analysis of _apg9_Δ indicates an import defect at an early step in the pathway. Wild-type (YW5-1B) and _apg9_Δ (CTD1) cells were grown in YPD (vegetative) or transferred to SD(−N) (starvation) for 3 h in the presence of PMSF (starvation) and examined by DIC (Nomarski) microscopy (Zeiss Axioplan). PMSF inhibits the degradation of autophagic bodies. Under starvation conditions, wild-type cells accumulate autophagic bodies when vesicle breakdown is blocked. However, under the same conditions, _apg9_Δ cells do not accumulate autophagic bodies, indicating that Apg9p is required for a step before vesicle fusion and release of the autophagic body into the vacuolar lumen. The _apg9_Δ strain transformed with the APG9 plasmid shows the same result as wild-type (data not shown). B, The apg9ts strain is tightly blocked for prAPI import at nonpermissive temperature. Wild-type (SEY6210) and _apg9_Δ (JKY007) cells transformed with the apg9ts centromeric plasmid were incubated at 24 and 38°C for 5 min, pulse-labeled for 10 min, and then subjected to nonradioactive chase reactions for the indicated times. Samples at each time point were immunoprecipitated with antiserum to API and resolved by SDS-PAGE as described in Materials and Methods. API-immunoprecipitated bands were quantified by a Molecular Dynamics STORM PhosphorImager and the results are presented in the graph. The percent mature API was determined by dividing the mature API value over the sum of the precursor and mature API values for each time point.
Figure 9
Vesicle accumulation test and kinetic analysis of prAPI import in the temperature conditional Apg9p mutant. A, Morphological analysis of _apg9_Δ indicates an import defect at an early step in the pathway. Wild-type (YW5-1B) and _apg9_Δ (CTD1) cells were grown in YPD (vegetative) or transferred to SD(−N) (starvation) for 3 h in the presence of PMSF (starvation) and examined by DIC (Nomarski) microscopy (Zeiss Axioplan). PMSF inhibits the degradation of autophagic bodies. Under starvation conditions, wild-type cells accumulate autophagic bodies when vesicle breakdown is blocked. However, under the same conditions, _apg9_Δ cells do not accumulate autophagic bodies, indicating that Apg9p is required for a step before vesicle fusion and release of the autophagic body into the vacuolar lumen. The _apg9_Δ strain transformed with the APG9 plasmid shows the same result as wild-type (data not shown). B, The apg9ts strain is tightly blocked for prAPI import at nonpermissive temperature. Wild-type (SEY6210) and _apg9_Δ (JKY007) cells transformed with the apg9ts centromeric plasmid were incubated at 24 and 38°C for 5 min, pulse-labeled for 10 min, and then subjected to nonradioactive chase reactions for the indicated times. Samples at each time point were immunoprecipitated with antiserum to API and resolved by SDS-PAGE as described in Materials and Methods. API-immunoprecipitated bands were quantified by a Molecular Dynamics STORM PhosphorImager and the results are presented in the graph. The percent mature API was determined by dividing the mature API value over the sum of the precursor and mature API values for each time point.
Figure 10
Analysis of the apg9ts strain indicates Apg9p is directly required for the vesicle formation step. A, The nature of prAPI binding and pelleting in the apg9ts strain. Spheroplasts of apg9ts were labeled for 10 min and chased for 30 min at 38°C. The spheroplasts were osmotically lysed in a buffer containing no salt (20 mM Pipes, pH 6.8) or a physiological concentration of salt (100 mM KOAc, 50 mM KCl, 5 mM MgCl2, 20 mM Pipes, pH 6.8) and pelleted at 5,000 g. All samples were immunoprecipitated with antiserum to API and the cytosolic marker PGK. apg9ts retains the salt dependent binding and pelleting of prAPI. B, prAPI in the apg9ts strain accumulates in both a membrane-associated state and a large pelletable complex. Spheroplasts of apg9ts and _ypt7_Δ were labeled for 10 min and chased for 30 min at 38°C. The labeled spheroplasts were then osmotically lysed and separated into supernatant (S) and pellet (P) fractions by centrifugation at 5,000 g for 5 min. An aliquot was removed for the total lysate control (T). The pellet fraction (P) was resuspended in 15% Ficoll-400 in gradient buffer (20 mM Pipes, 5 mM MgCl2, complete EDTA-free protease inhibitor cocktail) in the presence or absence of Triton X-100 and overlaid with 13 and 2% Ficoll-400 in gradient buffer. The step gradients were centrifuged at 13,000 g for 10 min. Membrane-containing float (F), nonfloat (NF), and pellet (P2) fractions were immunoprecipitated with antiserum to API as described in Materials and Methods. The position of prAPI is indicated. C, prAPI in the apg9ts strain is protease accessible. Spheroplasts isolated from apg9ts and _ypt7_Δ cells were pulse-labeled for 10 min and chased for 30 min at 38°C. The labeled spheroplasts were then osmotically lysed and separated into low-speed supernatant (S) and pellet (P) fractions after a 5,000 g centrifugation step. The pellet fractions were subjected to protease treatment in the absence or presence of 0.2% Triton X-100 as described in Materials and Methods. The resulting samples were immunoprecipitated with antiserum to API and PGK. The immunoprecipitated bands were quantified by a Molecular Dynamics STORM PhosphorImager. The percent protease-protected prAPI was determined by dividing the value for prAPI in the protease-treated sample by the value for total prAPI before protease treatment. The percent PGK was determined by dividing the supernatant (S) or pellet (P) value over the sum of the S and P values.
Figure 10
Analysis of the apg9ts strain indicates Apg9p is directly required for the vesicle formation step. A, The nature of prAPI binding and pelleting in the apg9ts strain. Spheroplasts of apg9ts were labeled for 10 min and chased for 30 min at 38°C. The spheroplasts were osmotically lysed in a buffer containing no salt (20 mM Pipes, pH 6.8) or a physiological concentration of salt (100 mM KOAc, 50 mM KCl, 5 mM MgCl2, 20 mM Pipes, pH 6.8) and pelleted at 5,000 g. All samples were immunoprecipitated with antiserum to API and the cytosolic marker PGK. apg9ts retains the salt dependent binding and pelleting of prAPI. B, prAPI in the apg9ts strain accumulates in both a membrane-associated state and a large pelletable complex. Spheroplasts of apg9ts and _ypt7_Δ were labeled for 10 min and chased for 30 min at 38°C. The labeled spheroplasts were then osmotically lysed and separated into supernatant (S) and pellet (P) fractions by centrifugation at 5,000 g for 5 min. An aliquot was removed for the total lysate control (T). The pellet fraction (P) was resuspended in 15% Ficoll-400 in gradient buffer (20 mM Pipes, 5 mM MgCl2, complete EDTA-free protease inhibitor cocktail) in the presence or absence of Triton X-100 and overlaid with 13 and 2% Ficoll-400 in gradient buffer. The step gradients were centrifuged at 13,000 g for 10 min. Membrane-containing float (F), nonfloat (NF), and pellet (P2) fractions were immunoprecipitated with antiserum to API as described in Materials and Methods. The position of prAPI is indicated. C, prAPI in the apg9ts strain is protease accessible. Spheroplasts isolated from apg9ts and _ypt7_Δ cells were pulse-labeled for 10 min and chased for 30 min at 38°C. The labeled spheroplasts were then osmotically lysed and separated into low-speed supernatant (S) and pellet (P) fractions after a 5,000 g centrifugation step. The pellet fractions were subjected to protease treatment in the absence or presence of 0.2% Triton X-100 as described in Materials and Methods. The resulting samples were immunoprecipitated with antiserum to API and PGK. The immunoprecipitated bands were quantified by a Molecular Dynamics STORM PhosphorImager. The percent protease-protected prAPI was determined by dividing the value for prAPI in the protease-treated sample by the value for total prAPI before protease treatment. The percent PGK was determined by dividing the supernatant (S) or pellet (P) value over the sum of the S and P values.
Similar articles
- Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole.
Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr, Klionsky DJ. Kim J, et al. J Cell Biol. 2001 Apr 16;153(2):381-96. doi: 10.1083/jcb.153.2.381. J Cell Biol. 2001. PMID: 11309418 Free PMC article. - The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways.
Huang WP, Scott SV, Kim J, Klionsky DJ. Huang WP, et al. J Biol Chem. 2000 Feb 25;275(8):5845-51. doi: 10.1074/jbc.275.8.5845. J Biol Chem. 2000. PMID: 10681575 - Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation.
Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Kim J, et al. J Biol Chem. 2002 Jan 4;277(1):763-73. doi: 10.1074/jbc.M109134200. Epub 2001 Oct 23. J Biol Chem. 2002. PMID: 11675395 Free PMC article. - Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae.
Khalfan WA, Klionsky DJ. Khalfan WA, et al. Curr Opin Cell Biol. 2002 Aug;14(4):468-75. doi: 10.1016/s0955-0674(02)00343-5. Curr Opin Cell Biol. 2002. PMID: 12383798 Review. - Autophagy in yeast: a review of the molecular machinery.
Huang WP, Klionsky DJ. Huang WP, et al. Cell Struct Funct. 2002 Dec;27(6):409-20. doi: 10.1247/csf.27.409. Cell Struct Funct. 2002. PMID: 12576634 Review.
Cited by
- The Knowns and Unknowns of Membrane Features and Changes During Autophagosome-Lysosome/Vacuole Fusion.
Liu J, Ma H, Wu Z, Ji Y, Liang Y. Liu J, et al. Int J Mol Sci. 2024 Oct 17;25(20):11160. doi: 10.3390/ijms252011160. Int J Mol Sci. 2024. PMID: 39456939 Free PMC article. Review. - Absence of ATG9A and synaptophysin demixing on Rab5 mutation-induced giant endosomes.
Choi J, Wu Y, Park D. Choi J, et al. Mol Brain. 2024 Sep 2;17(1):63. doi: 10.1186/s13041-024-01132-3. Mol Brain. 2024. PMID: 39223639 Free PMC article. - An APEX2-based proximity-dependent biotinylation assay with temporal specificity to study protein interactions during autophagy in the yeast Saccharomyces cerevisiae.
Filali-Mouncef Y, Leytens A, Vargas Duarte P, Zampieri M, Dengjel J, Reggiori F. Filali-Mouncef Y, et al. Autophagy. 2024 Oct;20(10):2323-2337. doi: 10.1080/15548627.2024.2366749. Epub 2024 Jul 3. Autophagy. 2024. PMID: 38958087 Free PMC article. - Tepsin binds LC3B to promote ATG9A trafficking and delivery.
Wallace NS, Gadbery JE, Cohen CI, Kendall AK, Jackson LP. Wallace NS, et al. Mol Biol Cell. 2024 Apr 1;35(4):ar56. doi: 10.1091/mbc.E23-09-0359-T. Epub 2024 Feb 21. Mol Biol Cell. 2024. PMID: 38381558 Free PMC article. - Role of autophagy in cancer-associated fibroblast activation, signaling and metabolic reprograming.
Sari D, Gozuacik D, Akkoc Y. Sari D, et al. Front Cell Dev Biol. 2024 Jan 3;11:1274682. doi: 10.3389/fcell.2023.1274682. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38234683 Free PMC article. Review.
References
- Baba M., Osumi M., Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases