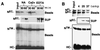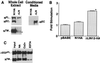Calcium depletion dissociates and activates heterodimeric notch receptors - PubMed (original) (raw)
Calcium depletion dissociates and activates heterodimeric notch receptors
M D Rand et al. Mol Cell Biol. 2000 Mar.
Abstract
Notch receptors participate in a highly conserved signaling pathway that regulates morphogenesis in multicellular animals. Maturation of Notch receptors requires the proteolytic cleavage of a single precursor polypeptide to produce a heterodimer composed of a ligand-binding extracellular domain (N(EC)) and a single-pass transmembrane signaling domain (N(TM)). Notch signaling has been correlated with additional ligand-induced proteolytic cleavages, as well as with nuclear translocation of the intracellular portion of N(TM) (N(ICD)). In the current work, we show that the N(EC) and N(TM) subunits of Drosophila Notch and human Notch1 (hN1) interact noncovalently. N(EC)-N(TM) interaction was disrupted by 0.1% sodium dodecyl sulfate or divalent cation chelators such as EDTA, and stabilized by millimolar Ca(2+). Deletion of the Ca(2+)-binding Lin12-Notch (LN) repeats from the N(EC) subunit resulted in spontaneous shedding of N(EC) into conditioned medium, implying that the LN repeats are important in maintaining the interaction of N(EC) and N(TM). The functional consequences of EDTA-induced N(EC) dissociation were studied by using hN1-expressing NIH 3T3 cells. Treatment of these cells for 10 to 15 min with 0.5 to 10 mM EDTA resulted in the rapid shedding of N(EC), the transient appearance of a polypeptide of the expected size of N(ICD), increased intranuclear anti-Notch1 staining, and the transient activation of an Notch-sensitive reporter gene. EDTA treatment of HeLa cells expressing endogenous Notch1 also stimulated reporter gene activity to a degree equivalent to that resulting from exposure of the cells to the ligand Delta1. These findings indicate that receptor activation can occur as a consequence of N(EC) dissociation, which relieves inhibition of the intrinsically active N(TM) subunit.
Figures
FIG. 1
Predicted structure of engineered Notch polypeptides. cDNAs encoding Drosophila Notch and various forms of human Notch1 were assembled and expressed in Drosophila S2 and mammalian cell lines, respectively, as described in Materials and Methods. dN, full-length Drosophila Notch; NEC, Notch extracellular subunit; NTM, Notch transmembrane subunit; L, leader peptide; HA, hemagglutinin tag; C, conserved cysteine residues; P, PEST sequence; N1HA, HA epitope-tagged human Notch 1; ΔEGF, a form of hN1 lacking all 36 EGF repeats; ΔLN-HA, a form of hN1 with the three LN module repeats deleted; NTM, a form of hN1 with an amino terminus 69 amino acids external to the transmembrane domain.
FIG. 2
Noncovalent association of Notch receptor subunits. The processed (NEC and NTM) and unprocessed (NFL) forms of Notch expressed in various cell lines were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) in reducing and nonreducing conditions and detected by Western blotting. (A) Proteins in whole-cell extracts (input) prepared from 106 N1HA cells with solutions containing either 1% NP-40 or 0.1% SDS were immunoprecipitated with anti-HA (αHA) or anti-Notch intracellular domain (αIC) antibody as indicated above each lane. After electrophoresis in SDS–6% polyacrylamide gels, Western blotting was done with anti-HA (in the top half of the panel) to detect the NFL and NEC and with anti-IC (in the lower half of the panel) to detect NTM. In the remaining panels, equivalent volumes of whole-cell extract prepared from N1HA (B), Jurkat (C), or Drosophila N-S2 (D) cells were solubilized in SDS loading buffer in the presence or absence of β-mercaptoethanol (ME), resolved by SDS-PAGE, and analyzed by Western blotting. N1HA and Jurkat cell blots were stained with anti-HA or anti-IC, and N-S2 blots were stained with antibodies against Notch intracellular (αIC, clone C17.9C6) or extracellular (αEC, clone F461.3B) domains, as indicated.
FIG. 3
EDTA-induced dissociation of NEC from NTM. The effect of EDTA on NEC-NTM stability was examined by using immunoprecipitates prepared from hN1HA cell lysates. (A) NEC-NTM immunocomplexes were prepared from a whole-cell extract of 2 × 106 N1HA cells with anti-IC on protein A-Sepharose beads. The beads were then divided into seven aliquots which were either held on ice (input) or incubated in TBS alone (NA), TBS containing 2 mM CaCl2 (Ca2+), or TBS containing 10 mM EDTA (EDTA) at 25°C for the times indicated. After the beads were pelleted, the resulting supernatants were incubated with anti-HA and protein A-beads to immunoprecipitate any released NEC. Immunoprecipitated proteins were analyzed by SDS-PAGE in 6% gels followed by Western blotting, using anti-HA to detect NEC (top and middle panels) and anti-IC to detect the NTM (bottom panel). HC, immunoglobulin H heavy chain. (B) To investigate the temperature dependence of EDTA-mediated dissociation of NEC, NEC-NTM immunocomplexes were prepared from a whole-cell lysate of 106 N1HA cells with anti-IC and protein A-beads. The beads were then divided into four aliquots that were either held on ice (input) or incubated for 15 min in TBS containing 5 mM EDTA at 4, 25, or 37°C. Beads and supernatants were then processed and analyzed as described for panel A.
FIG. 4
EDTA-induced dissociation of NEC from cells. (A) A total of 2 × 106 N-S2 cells were incubated for 30 min at 25°C in TBS buffer containing 5 mM CaCl2 (Ca2+), no additional ions (NA), or 5 mM EDTA. Polypeptides within the conditioned media were then resolved by SDS-PAGE on 7% gels and transferred to nitrocellulose. A Western blot is shown that is stained with antibody F461.3B directed against the dN extracellular domain. (B) Kc cells (107) were incubated for various times at 25°C in TBS containing 0.5 mM EDTA. In one experiment, cells were incubated for 60 min in TBS containing 0.5 mM EDTA and a cocktail of protease inhibitors (PI) consisting of 1 mM Preflabloc (Roche), 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml. (C) The effect of divalent metal ion chelation on the depletion of NEC from N-S2 cells was examined by a quantitative aggregation assay (see Materials and Methods). N-S2 cells were pretreated for 30 min at 25°C in 2 mM EDTA (■) or 5 mM CaCl2 (⧫). A molar excess of CaCl2 (5 mM) was then added to the EDTA-treated cells, which were incubated at additional 5 min. Dl-S2 cells were then added to Ca2+- or EDTA pretreated cells at 25°C. Aggregation was monitored by the change in transmitted light at 320 nm. (D) The concentration dependence of EDTA-mediated NEC shedding was investigated by incubating ∼2 × 105 N1HA cells for 15 min at 37°C in HBS containing the indicated concentrations of CaCl2 and/or EDTA. Released NEC was precipitated from the conditioned media with anti-HA on protein A-beads, resolved by SDS-PAGE in a 6% gel, and detected on a Western blot stained with anti-HA. (E) The time course of NEC dissociation from N1HA cells was determined by incubating ∼2 × 105 cells at 37°C in HBS containing either 2 mM CaCl2 (C) for 15 min or 0.5 mM EDTA for the times indicated. (F) The temperature dependence of NEC dissociation from N1HA cells was determined by incubating ∼2 × 105 cells in TBS containing 0.5 mM EDTA for 15 min at the temperatures indicated. In panels E and F, the release of NEC into conditioned media was analyzed as described for panel D. Western blots stained with anti-HA are shown.
FIG. 5
Requirement of LN repeats for NEC-NTM association. (A) Association of NEC and NTM subunits was assessed in NIH 3T3 cells expressing N1HA or a form of N1HA bearing a deletion that removes the coding sequence for all three LN modules (ΔLN; see Fig. 1). Whole-cell extracts (50 μg of total protein) from equivalent numbers of N1HA and ΔLN cells were analyzed on a divided Western blot stained with anti-HA (upper left panel) and anti-IC (lower left panel). The asterisk denotes a polypeptide of slightly smaller size than NTM that was observed only in ΔLN extracts; on shorter exposures, only the upper band was present in N1HA cell extracts. Media (∼8 ml) conditioned by ∼1 × 106 to 2 × 106 N1HA or ΔLN cells for 24 h was analyzed by immunoprecipitation with anti-HA to detect released NEC and ΔLNNEC (right panel). A representative Western blot is shown that is stained with anti-HA. (B) To determine the effect of various Notch1 polypeptides on CBF1-dependent signaling, equivalent numbers of pBABE control, N1HA, and ΔLN cells growing in six-well dishes were cotransfected in four independent experiments with a CBF1-sensitive firefly luciferase plasmid (HES-AB) and a Renilla luciferase control plasmid. Cell lysates prepared 36 h posttransfection were analyzed by dual luciferase assay. The fold stimulation was calculated as the ratio of normalized firefly luciferase activities to the mean activity in pBABE control lysates. Error bars indicate +1 standard error of the mean. The asterisk denotes differences from both pBABE and N1HA at P < 0.05. (C) ΔEGF, a cDNA encoding a form of hN1 bearing an extracellular-domain HA tag and a deletion removing the coding sequence for all 36 EGF repeats (see Fig. 1), was transiently expressed in dishes containing equivalent numbers (∼1 × 105) 293A cells. Processed and unprocessed ΔEGF polypeptides were immunoprecipitated from whole-cell extracts with anti-HA antibody on protein A-beads. Polypeptides retained on protein A-beads after incubation in TBS with either no addition (NA), 2.5 mM CaCl2 (Ca2+), or 10 mM EDTA for 30 min at 20°C were detected on a Western blot stained with anti-IC.
FIG. 6
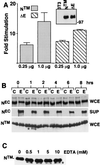
EDTA-induced NTM processing and nuclear translocation. (A) To demonstrate the intrinsic signaling activity of the NTM subunit, NIH 3T3 cells were cotransfected in triplicate with HES-AB luciferase reporter plasmid, Renilla luciferase control plasmid, and the indicated amounts of pcDNA3 plasmids containing either no cDNA insert, a cDNA insert encoding NTM, or a cDNA insert encoding ΔE. Firefly luciferase activities measured in cell lysates prepared 48 h posttransfection were normalized by using the corresponding internal Renilla luciferase control activities. The fold stimulation values were calculated as the ratio of individual normalized mean HES-AB-luciferase activities to the mean activity in control lysates prepared from cells transfected with empty pcDNA3 vector. In parallel experiments, extracts were prepared from NIH 3T3 cells transfected with either pcDNA3, pcDNA3-NTM, or pcDNA3-ΔE and the Renilla control plasmid. A Western blot normalized for differences in transfection efficiency (based on luciferase levels) was prepared and stained with rabbit anti-IC (inset). (B) ∼105 N1HA cells were treated for 15 min with HBS containing 2.5 mM CaCl2 (C) or 10 mM EDTA (E) at 37°C. The HBS-conditioned medium was harvested (SUP) and analyzed for release of NEC by preparation of immunoprecipitates with anti-HA, while cells were changed back to complete medium (D10) and allowed to recover for up to 8 h. Whole-cell lysates (WCE) prepared at various time points and immunoprecipitates prepared from conditioned media were analyzed on Western blots stained with anti-HA (upper and middle panels) or anti-IC (lower panel). An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells 1 h after exposure to EDTA. (C) A total of 2 × 105 N1HA cells were treated for 15 min with HBS containing either no addition (0) or increasing concentrations of EDTA at 37°C and then allowed to recover in D10 for 1 h. Whole-cell lysates were prepared and analyzed on a Western blot stained with anti-IC. An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells exposed to EDTA. (D) N1HA cells growing on slides were treated with HBS containing 0.5 mM EDTA for 15 min at 37°C, allowed to recover in D10 for 1 h (panel 1), 2 h (panel 2), or 4 h (panel 4) and then fixed and stained with anti-IC and goat anti-rabbit secondary antibody linked to FITC. The immunolocalization of Notch1 polypeptides in cells treated with EDTA was compared to control cells treated with HBS containing 2.5 mM CaCl2 for 15 min at 37°C followed by recovery in D10 for 1 h (panel C).
FIG. 6
EDTA-induced NTM processing and nuclear translocation. (A) To demonstrate the intrinsic signaling activity of the NTM subunit, NIH 3T3 cells were cotransfected in triplicate with HES-AB luciferase reporter plasmid, Renilla luciferase control plasmid, and the indicated amounts of pcDNA3 plasmids containing either no cDNA insert, a cDNA insert encoding NTM, or a cDNA insert encoding ΔE. Firefly luciferase activities measured in cell lysates prepared 48 h posttransfection were normalized by using the corresponding internal Renilla luciferase control activities. The fold stimulation values were calculated as the ratio of individual normalized mean HES-AB-luciferase activities to the mean activity in control lysates prepared from cells transfected with empty pcDNA3 vector. In parallel experiments, extracts were prepared from NIH 3T3 cells transfected with either pcDNA3, pcDNA3-NTM, or pcDNA3-ΔE and the Renilla control plasmid. A Western blot normalized for differences in transfection efficiency (based on luciferase levels) was prepared and stained with rabbit anti-IC (inset). (B) ∼105 N1HA cells were treated for 15 min with HBS containing 2.5 mM CaCl2 (C) or 10 mM EDTA (E) at 37°C. The HBS-conditioned medium was harvested (SUP) and analyzed for release of NEC by preparation of immunoprecipitates with anti-HA, while cells were changed back to complete medium (D10) and allowed to recover for up to 8 h. Whole-cell lysates (WCE) prepared at various time points and immunoprecipitates prepared from conditioned media were analyzed on Western blots stained with anti-HA (upper and middle panels) or anti-IC (lower panel). An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells 1 h after exposure to EDTA. (C) A total of 2 × 105 N1HA cells were treated for 15 min with HBS containing either no addition (0) or increasing concentrations of EDTA at 37°C and then allowed to recover in D10 for 1 h. Whole-cell lysates were prepared and analyzed on a Western blot stained with anti-IC. An asterisk denotes a novel anti-IC-cross-reactive polypeptide that appeared in lysates prepared from cells exposed to EDTA. (D) N1HA cells growing on slides were treated with HBS containing 0.5 mM EDTA for 15 min at 37°C, allowed to recover in D10 for 1 h (panel 1), 2 h (panel 2), or 4 h (panel 4) and then fixed and stained with anti-IC and goat anti-rabbit secondary antibody linked to FITC. The immunolocalization of Notch1 polypeptides in cells treated with EDTA was compared to control cells treated with HBS containing 2.5 mM CaCl2 for 15 min at 37°C followed by recovery in D10 for 1 h (panel C).
FIG. 7
EDTA-induced activation of CBF1-dependent transcription. (A) At 24 h after transfection with the HES-AB luciferase plasmid and a Renilla luciferase control plasmid, ∼2.5 × 104 N1HA cells were treated with HBS containing 2.5 mM CaCl2 (Ca++) or 10 mM EDTA for 15 min at 37°C. Cell lysates were subsequently prepared in triplicate after various periods of additional incubation in complete medium (D10). Firefly luciferase activities in lysates were normalized using the corresponding internal Renilla luciferase control activities, which were not affected by EDTA or CaCl2 treatment (not shown). The fold stimulation values were calculated as the ratio of individual normalized mean HES-AB luciferase activities to the mean activity in control lysates prepared from cells treated with HBS plus CaCl2 for 15 min and then immediately harvested. An asterisk denotes time points at which the mean firefly luciferase fold stimulation differs significantly from the corresponding CaCl2 control (P < 0.05). (B) N1HA cells were transfected as in panel A with the HES-AB-luciferase (HES) plasmid or the HES-ΔAB-luciferase plasmid (mHES) lacking functional CBF1 binding sites, plus a Renilla luciferase control plasmid. At 24 h posttransfection, cells were treated with 10 mM EDTA as in panel A and then allowed to recover for various times in D10. Normalized firefly luciferase activities were determined in cell lysates prepared in triplicate at each time point. The fold stimulation values were calculated as the ratio of individual normalized mean firefly luciferase activities to the mean activities of extracts prepared at 0 min. An asterisk denotes time points at which the fold stimulation of HES-AB luciferase activities differ significantly from the corresponding mHES control (P < 0.05). (C) N1HA cells were transfected as in panel A with the HES-AB-luciferase plasmid and a Renilla luciferase control plasmid. At 24 h posttransfection, cells were treated with HBS containing the concentrations of CaCl2 and/or EDTA indicated for 15 min at 37°C and then incubated an additional 6 h in D10 medium. Normalized mean luciferase activities were determined in cell lysates prepared in triplicate for each treatment. The fold stimulation values were calculated as the ratio of individual normalized mean firefly luciferase activities to the mean activity of lysates prepared after treatment with 2 mM CaCl2. An asterisk denotes the mean firefly luciferase activities that differ significantly from that of the 2 mM CaCl2 control (P < 0.05). (D) N1HA and pBABE control cells were transfected as in panel A with the HES-AB firefly luciferase plasmid and a Renilla luciferase plasmid. At 24 h posttransfection, cells were treated with HBS containing 2.5 mM CaCl2 or 10 mM EDTA for 15 min or 30 min at 37°C. After 6 h of recovery in D10, cell lysates were prepared. The fold stimulation values were calculated as the ratio of normalized mean firefly luciferase activities in EDTA-treated cell lysates to the mean activity in the corresponding CaCl2-treated cell lysates. An asterisk denotes the mean levels of firefly luciferase activity that differ significantly from the corresponding 3T3-pBABE control (P < 0.05). To detect expression of Notch2 NTM, a Western blot (inset) containing extracts from 3T3-pBABE cells (3T3; 50 μg of protein loaded) and was stained with the Notch2-specific rat monoclonal antibody bHN6 (46). An adjacent lane containing an extract of 293A cells transiently transfected with pcDNA3-ICN2 (5 μg of protein loaded) served as a positive control and demonstrated that the cross-reactive polypeptide in 3T3-pBABE cells was slightly larger in size than the intracellular domain of Notch2. (E) 3T3-pBABE cells were transfected as in panel A with the HES-AB firefly luciferase plasmid, a Renilla luciferase plasmid, and either pcDNA3 (2 μg) or pCMV-N3IC (2 μg). At 24 h posttransfection, cells transfected with pcDNA3 (control) or pCMV-N3IC (N3IC) were treated in triplicate with HBS containing 2.5 mM CaCl2 or 0.5 mM EDTA for 15 min at 37°C. After 6 h of recovery in D10, cell lysates were prepared. The fold stimulation values were calculated as the ratio of normalized mean firefly luciferase activities in EDTA-treated cell lysates to the mean activity in the corresponding CaCl2-treated cell lysates. An asterisk denotes the mean levels of firefly luciferase activity that differ significantly from the corresponding pBABE control (P < 0.05). (F) HeLa cells (seeded at 105 cells and grown for 2 days) were cotransfected with 0.06 μg of CMV-βGal plasmid and either 0.6 μg of HES-AB-luciferase or 0.6 μg of mHES-luciferase reporter. At 24 h after transfection, control QT6 and Delta-expressing QT6Dl cells (4 × 105) were added to the HeLa cells and maintained in coculture for 24 h. In separate wells, HeLa cells were also exposed to D10 media (control), PBS, or PBS containing 0.5 mM EDTA for 15 min at 37°C and then incubated an additional 6 h in D10 prior to determination of the luciferase activities. All treatments were performed in duplicate and luciferase activities were normalized by using the β-galactosidase activities. The fold stimulation values represent the ratio between individual mean normalized luciferase activities and the mean normalized activity produced by treatment with complete media alone, except for the QT6Dl-treated samples, which are compared to QT6 samples. The data represent the means of three independent experiments. Notch1 and Notch2 polypeptides in HeLa cell extracts were detected on Western blots (inset) stained with rat monoclonal anti-Notch1 (clone bTan20) or anti-Notch2 (clone bHN6). The error bars in panels A to F correspond to +1 standard error of the mean.
Similar articles
- Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats.
Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Sanchez-Irizarry C, et al. Mol Cell Biol. 2004 Nov;24(21):9265-73. doi: 10.1128/MCB.24.21.9265-9273.2004. Mol Cell Biol. 2004. PMID: 15485896 Free PMC article. - Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1.
Vardar D, North CL, Sanchez-Irizarry C, Aster JC, Blacklow SC. Vardar D, et al. Biochemistry. 2003 Jun 17;42(23):7061-7. doi: 10.1021/bi034156y. Biochemistry. 2003. PMID: 12795601 - Distinct roles of EGF repeats for the Notch signaling system.
Sakamoto K, Chao WS, Katsube K, Yamaguchi A. Sakamoto K, et al. Exp Cell Res. 2005 Jan 15;302(2):281-91. doi: 10.1016/j.yexcr.2004.09.016. Exp Cell Res. 2005. PMID: 15561108 - The role of notch in modeling and maintaining the vasculature.
Karsan A. Karsan A. Can J Physiol Pharmacol. 2005 Jan;83(1):14-23. doi: 10.1139/y04-125. Can J Physiol Pharmacol. 2005. PMID: 15759046 Review. - Structural conservation of Notch receptors and ligands.
Fleming RJ. Fleming RJ. Semin Cell Dev Biol. 1998 Dec;9(6):599-607. doi: 10.1006/scdb.1998.0260. Semin Cell Dev Biol. 1998. PMID: 9918871 Review.
Cited by
- The notch ligand jagged-1 represents a novel growth factor of human hematopoietic stem cells.
Karanu FN, Murdoch B, Gallacher L, Wu DM, Koremoto M, Sakano S, Bhatia M. Karanu FN, et al. J Exp Med. 2000 Nov 6;192(9):1365-72. doi: 10.1084/jem.192.9.1365. J Exp Med. 2000. PMID: 11067884 Free PMC article. - Spatial segregation of gamma-secretase and substrates in distinct membrane domains.
Vetrivel KS, Cheng H, Kim SH, Chen Y, Barnes NY, Parent AT, Sisodia SS, Thinakaran G. Vetrivel KS, et al. J Biol Chem. 2005 Jul 8;280(27):25892-900. doi: 10.1074/jbc.M503570200. Epub 2005 May 10. J Biol Chem. 2005. PMID: 15886206 Free PMC article. - Overexpression of miRNA-25-3p inhibits Notch1 signaling and TGF-β-induced collagen expression in hepatic stellate cells.
Genz B, Coleman MA, Irvine KM, Kutasovic JR, Miranda M, Gratte FD, Tirnitz-Parker JEE, Olynyk JK, Calvopina DA, Weis A, Cloonan N, Robinson H, Hill MM, Al-Ejeh F, Ramm GA. Genz B, et al. Sci Rep. 2019 Jun 12;9(1):8541. doi: 10.1038/s41598-019-44865-1. Sci Rep. 2019. PMID: 31189969 Free PMC article. - AIP4/Itch regulates Notch receptor degradation in the absence of ligand.
Chastagner P, Israël A, Brou C. Chastagner P, et al. PLoS One. 2008 Jul 16;3(7):e2735. doi: 10.1371/journal.pone.0002735. PLoS One. 2008. PMID: 18628966 Free PMC article. - Ligand-Independent Mechanisms of Notch Activity.
Palmer WH, Deng WM. Palmer WH, et al. Trends Cell Biol. 2015 Nov;25(11):697-707. doi: 10.1016/j.tcb.2015.07.010. Epub 2015 Oct 1. Trends Cell Biol. 2015. PMID: 26437585 Free PMC article. Review.
References
- Ahmad I, Zagouras P, Artavanis-Tsakonis S. Involvement of Notch-1 in mammalian retinal neurogenesis: association of Notch-1 activity with both immature and terminally differentiated cells. Mech Dev. 1995;53:73–85. - PubMed
- Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
- Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. - PubMed
- Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
- Aster J C, Simms W B, Zavala-Ruiz Z, Patriub V, North C L, Blacklow S C. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry. 1999;38:4736–4742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA66849/CA/NCI NIH HHS/United States
- R37 NS026084/NS/NINDS NIH HHS/United States
- CA62450/CA/NCI NIH HHS/United States
- CA82308/CA/NCI NIH HHS/United States
- R01 CA062450/CA/NCI NIH HHS/United States
- R01 NS026084/NS/NINDS NIH HHS/United States
- R01 CA082308/CA/NCI NIH HHS/United States
- F32 NS010735/NS/NINDS NIH HHS/United States
- R01 HL061001/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous