High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR - PubMed (original) (raw)
High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR
S Germer et al. Genome Res. 2000 Feb.
Abstract
We have developed an accurate, yet inexpensive and high-throughput, method for determining the allele frequency of biallelic polymorphisms in pools of DNA samples. The assay combines kinetic (real-time quantitative) PCR with allele-specific amplification and requires no post-PCR processing. The relative amounts of each allele in a sample are quantified. This is performed by dividing equal aliquots of the pooled DNA between two separate PCR reactions, each of which contains a primer pair specific to one or the other allelic SNP variant. For pools with equal amounts of the two alleles, the two amplifications should reach a detectable level of fluorescence at the same cycle number. For pools that contain unequal ratios of the two alleles, the difference in cycle number between the two amplification reactions can be used to calculate the relative allele amounts. We demonstrate the accuracy and reliability of the assay on samples with known predetermined SNP allele frequencies from 5% to 95%, including pools of both human and mouse DNAs using eight different SNPs altogether. The accuracy of measuring known allele frequencies is very high, with the strength of correlation between measured and known frequencies having an r(2) = 0.997. The loss of sensitivity as a result of measurement error is typically minimal, compared with that due to sampling error alone, for population samples up to 1000. We believe that by providing a means for SNP genotyping up to thousands of samples simultaneously, inexpensively, and reproducibly, this method is a powerful strategy for detecting meaningful polymorphic differences in candidate gene association studies and genome-wide linkage disequilibrium scans.
Figures
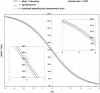
Figure 1
The basis of allele frequency measurement using kinetic PCR. Shown are amplification growth curves of PCR reactions performed for the ApoB71 polymorphism. A sample was constructed from two DNAs each homozygous for the different alleles of the ApoB71 SNP and contains 5% of allele 1. Equal aliquots of the pool (20 ng of DNA each) were put into PCRs containing either of the two allele-specific primer sets. Four replicate reactions were performed with each primer set (eight PCRs total). The relative allele frequency is determined on the basis of the Δ_Ct_ using equation 1 (see text and Fig. 2).
Figure 2
The relationship between Δ_Ct_ and allele frequency. The solid center line is a plot of equation 1 from the text. The flanking solid lines represent the expected uncertainty (1
s.d.
) in estimating the allele frequency based on sampling error alone (sample size = 1000). The broken lines represent the combined uncertainty of sampling and measurement error. The measurement error is based on an average error seen amongst the measurements taken in this paper and is that expected after averaging four replicate measurements. The insets compare the impact of measurement error at the middle and at the upper extreme of allele frequencies (the lower extreme should mirror exactly the upper).
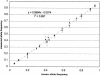
Figure 3
The accuracy of allele frequency measurement by kinetic PCR. Shown is a scatter-plot of all the measurements of allele frequency made in Tables 1, 2, and 3 comparing the known frequencies (determined by DNA concentration for Table 1 and by individual genotyping and allele counting for Tables 1 and 2) with the measured frequencies. The error bars represent one
s.d.
in the measurement. The diagonal line is that expected for complete concordance between known and measured values.
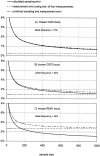
Figure 4
The impact of measurement error for three SNP assays. (A) Plotted as the solid line is the expected sampling error for this SNP given an allele frequency of 10% (see text) for sample sizes up to 1000. The upper broken line is the estimated combined sampling and measurement error for this assay based on Table 3 and using the average of four measurements. This measurement error alone is the lower broken line. (B) The same as A for the human CST5 locus (Table 2). (C) The same as A and B for the mouse REN1 SNP (Table 3).
Similar articles
- Quantitative determination of allele frequency in pooled DNA by using sequencing method.
Cao P, Wang QJ, Zhu XT, Zhou H, Li R, Wang WP. Cao P, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2011 Mar 1;879(7-8):527-32. doi: 10.1016/j.jchromb.2011.01.014. Epub 2011 Jan 15. J Chromatogr B Analyt Technol Biomed Life Sci. 2011. PMID: 21277843 - Microarray-based estimation of SNP allele-frequency in pooled DNA using the Langmuir kinetic model.
Yin BC, Li H, Ye BC. Yin BC, et al. BMC Genomics. 2008 Dec 16;9:605. doi: 10.1186/1471-2164-9-605. BMC Genomics. 2008. PMID: 19087310 Free PMC article. - Kinetic polymerase chain reaction on pooled DNA: a high-throughput, high-efficiency alternative in genetic epidemiological studies.
Chen J, Germer S, Higuchi R, Berkowitz G, Godbold J, Wetmur JG. Chen J, et al. Cancer Epidemiol Biomarkers Prev. 2002 Jan;11(1):131-6. Cancer Epidemiol Biomarkers Prev. 2002. PMID: 11815411 - Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools.
Norton N, Williams NM, Williams HJ, Spurlock G, Kirov G, Morris DW, Hoogendoorn B, Owen MJ, O'Donovan MC. Norton N, et al. Hum Genet. 2002 May;110(5):471-8. doi: 10.1007/s00439-002-0706-6. Epub 2002 Mar 23. Hum Genet. 2002. PMID: 12073018 - Approaches to allele frequency determination.
Kwok PY. Kwok PY. Pharmacogenomics. 2000 May;1(2):231-5. doi: 10.1517/14622416.1.2.231. Pharmacogenomics. 2000. PMID: 11256594 Review.
Cited by
- An association study between the transthyretin (TTR) gene and mental retardation.
Li J, Gao JJ, Zhang FC, Xing QH, Dang FL, Gao XC, Duan SW, Zheng ZJ, Qian XQ, Qin W, Li XW, Han YF, Li J, Feng GY, St Clair D, He L. Li J, et al. Eur Arch Psychiatry Clin Neurosci. 2006 Jun;256(4):230-5. doi: 10.1007/s00406-005-0630-6. Epub 2005 Dec 19. Eur Arch Psychiatry Clin Neurosci. 2006. PMID: 16362527 - Gene-to-gene interaction between sodium channel-related genes in determining the risk of antiepileptic drug resistance.
Jang SY, Kim MK, Lee KR, Park MS, Kim BC, Cho KH, Lee MC, Kim YS. Jang SY, et al. J Korean Med Sci. 2009 Feb;24(1):62-8. doi: 10.3346/jkms.2009.24.1.62. Epub 2009 Feb 28. J Korean Med Sci. 2009. PMID: 19270815 Free PMC article. - Whole-genome sequencing of Burkholderia pseudomallei isolates from an unusual melioidosis case identifies a polyclonal infection with the same multilocus sequence type.
Price EP, Sarovich DS, Viberg L, Mayo M, Kaestli M, Tuanyok A, Foster JT, Keim P, Pearson T, Currie BJ. Price EP, et al. J Clin Microbiol. 2015 Jan;53(1):282-6. doi: 10.1128/JCM.02560-14. Epub 2014 Oct 22. J Clin Microbiol. 2015. PMID: 25339397 Free PMC article. - One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes.
Sotlar K, Escribano L, Landt O, Möhrle S, Herrero S, Torrelo A, Lass U, Horny HP, Bültmann B. Sotlar K, et al. Am J Pathol. 2003 Mar;162(3):737-46. doi: 10.1016/S0002-9440(10)63870-9. Am J Pathol. 2003. PMID: 12598308 Free PMC article. - An IL28B genotype-based clinical prediction model for treatment of chronic hepatitis C.
O'Brien TR, Everhart JE, Morgan TR, Lok AS, Chung RT, Shao Y, Shiffman ML, Dotrang M, Sninsky JJ, Bonkovsky HL, Pfeiffer RM; HALT-C Trial Group. O'Brien TR, et al. PLoS One. 2011;6(7):e20904. doi: 10.1371/journal.pone.0020904. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760886 Free PMC article. Clinical Trial.
References
- Balbin M, Freije JP, Abrahamson M, Velasco G, Grubb A, López-Otín C. A sequence variation in the human cystatin D gene resulting in an amino acid (Cys/Arg) polymorphism at the protein level. Hum Genet. 1993;90:668–669. - PubMed
- Beasley EM, Myers RM, Cox DR, Lazzaroni LC. Statistical refinement of primer design parameters. In: Innes MA, Gelfand DH, Sninsky JJ, editors. PCR applications: Protocols for functional genomics. San Diego, CA: Academic Press; 1999. pp. 55–73.
- Birch DE. Simplified hot start PCR. Nature. 1996;381:445–446. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources