Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK - PubMed (original) (raw)
. 2000 Feb 1;14(3):349-59.
Affiliations
- PMID: 10673506
- PMCID: PMC316364
Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK
S J Araújo et al. Genes Dev. 2000.
Abstract
During human nucleotide excision repair, damage is recognized, two incisions are made flanking a DNA lesion, and residues are replaced by repair synthesis. A set of proteins required for repair of most lesions is RPA, XPA, TFIIH, XPC-hHR23B, XPG, and ERCC1-XPF, but additional components have not been excluded. The most complex and difficult to analyze factor is TFIIH, which has a 6-subunit core (XPB, XPD, p44, p34, p52, p62) and a 3-subunit kinase (CAK). TFIIH has roles both in basal transcription initiation and in DNA repair, and several inherited human disorders are associated with mutations in TFIIH subunits. To identify the forms of TFIIH that can function in repair, recombinant XPA, RPA, XPC-hHR23B, XPG, and ERCC1-XPF were combined with TFIIH fractions purified from HeLa cells. Repair activity coeluted with the peak of TFIIH and with transcription activity. TFIIH from cells with XPB or XPD mutations was defective in supporting repair, whereas TFIIH from spinal muscular atrophy cells with a deletion of one p44 gene was active. Recombinant TFIIH also functioned in repair, both a 6- and a 9-subunit form containing CAK. The CAK kinase inhibitor H-8 improved repair efficiency, indicating that CAK can negatively regulate NER by phosphorylation. The 15 recombinant polypeptides define the minimal set of proteins required for dual incision of DNA containing a cisplatin adduct. Complete repair was achieved by including highly purified human DNA polymerase delta or epsilon, PCNA, RFC, and DNA ligase I in reaction mixtures, reconstituting adduct repair for the first time with recombinant incision factors and human replication proteins.
Figures
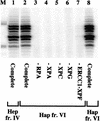
Figure 1
Purified human proteins for NER. (A) Incision factors (top line) and repair synthesis factors (bottom line). PCNA, Lig1, and all incision factors are recombinant polypeptides except for TFIIH Hap fr. VI. The XPA, RPA, XPG, PCNA, and Lig1 gels were stained with Coomassie blue and the others with silver. Asterisks represent identified subunits of human RFC, Pol δ, and Pol ε. (B) Recombinant TFIIH. Silver-stained gels (left) and immunoblots (right); equivalent amounts of rIIH6 or rIIH9 were loaded. The six core subunits are present in similar amounts in the two preparations, and the CAK subunits in rIIH9 appear to be roughly stoichiometric with the core subunits.
Figure 1
Purified human proteins for NER. (A) Incision factors (top line) and repair synthesis factors (bottom line). PCNA, Lig1, and all incision factors are recombinant polypeptides except for TFIIH Hap fr. VI. The XPA, RPA, XPG, PCNA, and Lig1 gels were stained with Coomassie blue and the others with silver. Asterisks represent identified subunits of human RFC, Pol δ, and Pol ε. (B) Recombinant TFIIH. Silver-stained gels (left) and immunoblots (right); equivalent amounts of rIIH6 or rIIH9 were loaded. The six core subunits are present in similar amounts in the two preparations, and the CAK subunits in rIIH9 appear to be roughly stoichiometric with the core subunits.
Figure 2
Transcription and NER activities of TFIIH copurify. (A) Scheme for TFIIH purification from HeLa cells; TFIIH Hep is the result of step IV, heparin–HPLC and TFIIH Hap is the result of step VI, hydroxyapatite–HPLC. (B) (Top panel) In vitro runoff transcription assays (Tirode et al. 1999), using 1 μl of each fraction; (middle panel) 5 μl of each step V fraction was separated by 10% SDS-PAGE and immunoblotted against the three indicated subunits of TFIIH; (bottom panel) 3 μl of each step V fraction was used in a nucleotide excision repair assay containing all the recombinant proteins needed for dual incision formation (except TFIIH); 1.5 μl of TFIIH Hep fr. IV was used where indicated. Only the area of the gel containing NER excision products is shown. (C) As in B, for step VI TFIIH fractions. (M) 35- and 27-nucleotide molecular weight markers from a labeled _Msp_I digest of pBR322.
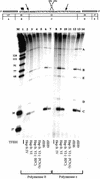
Figure 3
Dual incision with highly purified TFIIH from human cells. NER was performed in the presence of recombinant RPA, XPA, XPC–hHR23B, XPG, ERCC1–XPF, and TFIIH fr. IV or fr. VI as indicated. Repair factors were individually omitted as shown. Amounts of TFIIH used were 2 μl of Hap fr. VI (lanes 2_–_7), 1.5 μl of Hep fr. IV (lane 1), and 2 μl of Hap fr. VI from a different purification (lane 8); TFIIH Hap fr. VI is fraction 6 from Fig. 2_C_.
Figure 4
Dual incision reconstituted with recombinant proteins: Recombinant TFIIH is active in NER. Different amounts of rIIH6 and rIIH9 were used in a nucleotide excision repair assay containing all the recombinant proteins needed for dual incision formation, as indicated. The TFIIH Hep fr. IV lane used 1.5 μl of fraction, and the HeLa lane used 20 μg of whole cell extract protein.
Figure 5
Role of CAK in dual incision formation. (A) rIIH6 (2 μl) was tested in a dual incision assay together with increasing amounts of rCAK in the presence (lanes 6_–_9) or absence (lanes 1_–_5) of an ATP-regenerating system. The effect of H-8 in the presence of CAK was measured by adding the inhibitor to a final concentration of 100 μ
m
(lanes 3,7). Data were normalized to the amount of repair obtained in reaction mixtures containing TFIIH Hep fr. IV instead of rIIH6. (B) TFIIH Hep fr. IV (1.5 μl) was tested for dual incision formation in the presence (lanes 6_–_10) or absence (lanes 1_–_5) of the same ATP-regenerating system as in A; increasing amounts of H-8 kinase inhibitor were added as indicated. (C) TFIIH purified from HeLa cells without the CAK subunit [TFIIH (−CAK), 2 μl] was tested in a dual incision assay in the presence of ATP-regenerating system. TFIIH (−CAK) was prepared by disrupting TFIIH complex with buffer containing 1.2
m
KCl and depleting the CAK by immunopurification with a cdk7 antibody (Rossignol et al. 1997), and so is less concentrated than TFIIH Hep fr. IV. Increasing amounts of H-8 were added as indicated (lanes 4_–_6); 1 μl of rCAK was added to a reaction mixture in the absence of H-8 (lane 7). (D) TFIIH Hap fr. VI (1.5 μl) was tested in a dual incision assay in the presence of the ATP-regenerating system and increasing amounts of H-8 as indicated (lanes 3_–_5). In all panels 1.5 μl of Hep fr. IV was used. Quantification was done on a PhosphorImager with ImageQuant. The density of the bands corresponding to the 26- to 30-mer products was quantified and divided by the value corresponding to the reactions performed in the presence of TFIIH Hep fr. IV to give the relative counts plotted in the graph.
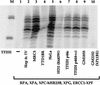
Figure 6
NER activity of TFIIH containing mutations in different subunits. TFIIH samples from repair-proficient cell lines (MRC5, HeLa, and GM1855, heterozygous normal parent of XP11BE), XP-B (GM2252, patient XP11BE), HD2 (XP-D mutation R683W), TTD-A (TTD1BR), SMA line RA (TFIIH p44c), and SMA line DJ [TFIIH p44(t+c)] were tested for dual incision activity in combination with the other recombinant core incision components as indicated. The TFIIH concentration of each sample was estimated by immunoblotting (Coin et al. 1999), and equivalent amounts of the different preparations were used in the incision assays.
Figure 7
Reconstitution of incision and repair synthesis with recombinant and human purified factors. (Top) Schematic representation of the DNA substrate containing a single 1,3 intrastrand d(GpTpG) cisplatin cross-link. The sizes of the fragments generated by _Bst_NI restriction digestion are shown underneath. Arrows indicate mapped positions of NER incisions. (Bottom) Reconstitution of repair synthesis on a plasmid containing a single cisplatin adduct. Different TFIIH preparations were used in reactions performed in the presence of Pol δ (lanes 1_–_7) or Pol ε (lanes 8_–_14); lanes 2 and 9 contain 1.5 μl of TFIIH Hep fr. IV, lanes 3_–_5 and 10_–_12 contain 3 μl of TFIIH Hap fr. VI, lanes 6 and 13 contain 3 μl of rIIH6, and lanes 7 and 14 contain 3 μl of rIIH9. Reactions in lanes 4 and 11 were done in the absence of RFC, and reactions in lanes 5 and 12 were done in the absence of PCNA.
Similar articles
- Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH.
Rossignol M, Kolb-Cheynel I, Egly JM. Rossignol M, et al. EMBO J. 1997 Apr 1;16(7):1628-37. doi: 10.1093/emboj/16.7.1628. EMBO J. 1997. PMID: 9130708 Free PMC article. - Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH.
Reardon JT, Ge H, Gibbs E, Sancar A, Hurwitz J, Pan ZQ. Reardon JT, et al. Proc Natl Acad Sci U S A. 1996 Jun 25;93(13):6482-7. doi: 10.1073/pnas.93.13.6482. Proc Natl Acad Sci U S A. 1996. PMID: 8692841 Free PMC article. - Mechanism of open complex and dual incision formation by human nucleotide excision repair factors.
Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Evans E, et al. EMBO J. 1997 Nov 3;16(21):6559-73. doi: 10.1093/emboj/16.21.6559. EMBO J. 1997. PMID: 9351836 Free PMC article. - XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.
Fuss JO, Tainer JA. Fuss JO, et al. DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review. - DNA damage recognition during nucleotide excision repair in mammalian cells.
Wood RD. Wood RD. Biochimie. 1999 Jan-Feb;81(1-2):39-44. doi: 10.1016/s0300-9084(99)80036-4. Biochimie. 1999. PMID: 10214908 Review.
Cited by
- New synthetic substrates of mammalian nucleotide excision repair system.
Evdokimov A, Petruseva I, Tsidulko A, Koroleva L, Serpokrylova I, Silnikov V, Lavrik O. Evdokimov A, et al. Nucleic Acids Res. 2013 Jul;41(12):e123. doi: 10.1093/nar/gkt301. Epub 2013 Apr 22. Nucleic Acids Res. 2013. PMID: 23609543 Free PMC article. - Detection of the Excised, Damage-containing Oligonucleotide Products of Nucleotide Excision Repair in Human Cells.
Song J, Kemp MG, Choi JH. Song J, et al. Photochem Photobiol. 2017 Jan;93(1):192-198. doi: 10.1111/php.12638. Epub 2016 Nov 3. Photochem Photobiol. 2017. PMID: 27634428 Free PMC article. Review. - Nucleotide excision repair functions in the removal of chromium-induced DNA damage in mammalian cells.
O'Brien TJ, Brooks BR, Patierno SR. O'Brien TJ, et al. Mol Cell Biochem. 2005 Nov;279(1-2):85-95. doi: 10.1007/s11010-005-8225-0. Mol Cell Biochem. 2005. PMID: 16283517 - Both XPD alleles contribute to the phenotype of compound heterozygote xeroderma pigmentosum patients.
Ueda T, Compe E, Catez P, Kraemer KH, Egly JM. Ueda T, et al. J Exp Med. 2009 Dec 21;206(13):3031-46. doi: 10.1084/jem.20091892. Epub 2009 Nov 23. J Exp Med. 2009. PMID: 19934020 Free PMC article. - BRCA1 can modulate RNA polymerase II carboxy-terminal domain phosphorylation levels.
Moisan A, Larochelle C, Guillemette B, Gaudreau L. Moisan A, et al. Mol Cell Biol. 2004 Aug;24(16):6947-56. doi: 10.1128/MCB.24.16.6947-6956.2004. Mol Cell Biol. 2004. PMID: 15282296 Free PMC article.
References
- Aboussekhra A, Biggerstaff M, Shivji MKK, Vilpo JA, Moncollin V, Podust VN, Protic' M, Hübscher U, Egly J-M, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
- Araújo SJ, Wood RD. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. - PubMed
- Biggerstaff M, Wood RD. Assay for nucleotide excision repair proteins using mammalian cell extracts and UV damaged plasmid DNA. In: Henderson DS, editor. DNA repair protocols: Eukaryotic systems. Totowa, NJ: Humana Press; 1999. pp. 357–372.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous