Survey of the fragile X syndrome CGG repeat and the short-tandem-repeat and single-nucleotide-polymorphism haplotypes in an African American population - PubMed (original) (raw)
Comparative Study
doi: 10.1086/302762.
C E Schwartz, K L Meadows, J L Newman, L F Taft, C Gunter, W T Brown, N J Carpenter, P N Howard-Peebles, K G Monaghan, S L Nolin, A L Reiss, G L Feldman, E M Rohlfs, S T Warren, S L Sherman
Affiliations
- PMID: 10677308
- PMCID: PMC1288101
- DOI: 10.1086/302762
Comparative Study
Survey of the fragile X syndrome CGG repeat and the short-tandem-repeat and single-nucleotide-polymorphism haplotypes in an African American population
D C Crawford et al. Am J Hum Genet. 2000 Feb.
Abstract
Previous studies have shown that specific short-tandem-repeat (STR) and single-nucleotide-polymorphism (SNP)-based haplotypes within and among unaffected and fragile X white populations are found to be associated with specific CGG-repeat patterns. It has been hypothesized that these associations result from different mutational mechanisms, possibly influenced by the CGG structure and/or cis-acting factors. Alternatively, haplotype associations may result from the long mutational history of increasing instability. To understand the basis of the mutational process, we examined the CGG-repeat size, three flanking STR markers (DXS548-FRAXAC1-FRAXAC2), and one SNP (ATL1) spanning 150 kb around the CGG repeat in unaffected (n=637) and fragile X (n=63) African American populations and compared them with unaffected (n=721) and fragile X (n=102) white populations. Several important differences were found between the two ethnic groups. First, in contrast to that seen in the white population, no associations were observed among the African American intermediate or "predisposed" alleles (41-60 repeats). Second, two previously undescribed haplotypes accounted for the majority of the African American fragile X population. Third, a putative "protective" haplotype was not found among African Americans, whereas it was found among whites. Fourth, in contrast to that seen in whites, the SNP ATL1 was in linkage equilibrium among African Americans, and it did not add new information to the STR haplotypes. These data indicate that the STR- and SNP-based haplotype associations identified in whites probably reflect the mutational history of the expansion, rather than a mutational mechanism or pathway.
Figures
Figure 1
Location and nomenclature of FRAXA interspersion patterns and STR- and SNP-based haplotypes. This figure was adapted from Eichler et al. (1996). We show here the position of the STR and SNP markers relative to the FRAXA CGG repeat. We also included the FRAXE GCC repeat, the STR marker DXS1691, and the recently described polymorphism within DXS548 (DXS548-P). The numbers above the marker represent the allele assignment and the numbers in parentheses represent the base-pair size, according to our protocol. The numbering systems and interspersion-pattern nomenclature are from Eichler et al. (1996) and Murray et al. (1996). We have extended the numbering system for DXS548 and FRAXAC2 to include the alleles not previously described in a white population. All STR-based haplotypes are constructed as follows: 5′-DXS548-P (if there is an insertion or deletion event)-DXS548-FRAXAC1-FRAXAC2-3′. Interspersion patterns are described as the number of consecutive CGGs followed by a plus sign (+) to designate the presence of an AGG interruption.
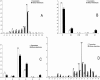
Figure 2
A, DXS548; B, DXS548-P; C, FRAXAC1; and D, FRAXAC2 allele distributions in whites (
_n_=721
; unshaded bars) and African Americans (
_n_=637
; shaded bars). Absolute numbers are reported above each bar for each allele of each locus in both populations.
Similar articles
- Genetic diversity of the fragile X syndrome gene (FMR1) in a large Sub-Saharan West African population.
Peprah EK, Allen EG, Williams SM, Woodard LM, Sherman SL. Peprah EK, et al. Ann Hum Genet. 2010 Jul;74(4):316-25. doi: 10.1111/j.1469-1809.2010.00582.x. Ann Hum Genet. 2010. PMID: 20597902 Free PMC article. - A new insight into fragile X syndrome among Basque population.
Peñagarikano O, Gil A, Télez M, Ortega B, Flores P, Veiga I, Peixoto A, Criado B, Arrieta I. Peñagarikano O, et al. Am J Med Genet A. 2004 Jul 30;128A(3):250-5. doi: 10.1002/ajmg.a.30116. Am J Med Genet A. 2004. PMID: 15216545 - [Molecular and genetic features of fragile X syndrome].
Jara L, Avendaño I, Aspillaga M, Blanco R. Jara L, et al. Rev Med Chil. 1996 Jul;124(7):865-72. Rev Med Chil. 1996. PMID: 9138377 Review. Spanish. - An assessment of screening strategies for fragile X syndrome in the UK.
Pembrey ME, Barnicoat AJ, Carmichael B, Bobrow M, Turner G. Pembrey ME, et al. Health Technol Assess. 2001;5(7):1-95. doi: 10.3310/hta5070. Health Technol Assess. 2001. PMID: 11262423 Review.
Cited by
- Expansion of an FMR1 grey-zone allele to a full mutation in two generations.
Fernandez-Carvajal I, Lopez Posadas B, Pan R, Raske C, Hagerman PJ, Tassone F. Fernandez-Carvajal I, et al. J Mol Diagn. 2009 Jul;11(4):306-10. doi: 10.2353/jmoldx.2009.080174. Epub 2009 Jun 12. J Mol Diagn. 2009. PMID: 19525339 Free PMC article. - The AGG interruption pattern within the CGG repeat of the FMR1 gene among Taiwanese population.
Chiu HH, Tseng YT, Hsiao HP, Hsiao HH. Chiu HH, et al. J Genet. 2008 Dec;87(3):275-7. doi: 10.1007/s12041-008-0043-1. J Genet. 2008. PMID: 19147914 No abstract available. - FMR1 haplotype analyses among Indians: a weak founder effect and other findings.
Sharma D, Gupta M, Thelma BK. Sharma D, et al. Hum Genet. 2003 Mar;112(3):262-71. doi: 10.1007/s00439-002-0872-6. Epub 2002 Dec 14. Hum Genet. 2003. PMID: 12596051 - Contraction of fully expanded FMR1 alleles to the normal range: predisposing haplotype or rare events?
Maia N, Loureiro JR, Oliveira B, Marques I, Santos R, Jorge P, Martins S. Maia N, et al. J Hum Genet. 2017 Feb;62(2):269-275. doi: 10.1038/jhg.2016.122. Epub 2016 Oct 27. J Hum Genet. 2017. PMID: 27784894 - Unique AGG Interruption in the CGG Repeats of the FMR1 Gene Exclusively Found in Asians Linked to a Specific SNP Haplotype.
Limprasert P, Thanakitgosate J, Jaruthamsophon K, Sripo T. Limprasert P, et al. Genet Res Int. 2016;2016:8319287. doi: 10.1155/2016/8319287. Epub 2016 Mar 2. Genet Res Int. 2016. PMID: 27042357 Free PMC article.
References
Electronic-Database Information
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for fragile X syndrome [MIM <309550>])
References
- Arrieta I, Gil A, Nunez T, Elez M, Artinez B, Criado B, Stao C (1999) Stability of the FMR1 CGG repeat in a Basque sample. Hum Biol 71:55–68 - PubMed
- Brown TC, Tarleton JC, Go RC, Longshore JW, Descartes M (1997) Instability of the FMR2 trinucleotide repeat region associated with expanded FMR1 alleles. Am J Med Genet 73:447–455 - PubMed
- Brown WT, Houck GEJ, Jeziorowska A, Levinson FN, Ding X, Dobkin C, Zhong N, et al (1993) Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA 270:1569–1575 [published erratum appears in JAMA 271:28] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HD035576/HD/NICHD NIH HHS/United States
- M01 RR000039/RR/NCRR NIH HHS/United States
- R01 HD029909/HD/NICHD NIH HHS/United States
- HD29909/HD/NICHD NIH HHS/United States
- HD35576/HD/NICHD NIH HHS/United States
- MO1-RR-00039/RR/NCRR NIH HHS/United States
- F31 HD008443/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical