Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis - PubMed (original) (raw)
Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis
N Kaminski et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The molecular mechanisms of pulmonary fibrosis are poorly understood. We have used oligonucleotide arrays to analyze the gene expression programs that underlie pulmonary fibrosis in response to bleomycin, a drug that causes lung inflammation and fibrosis, in two strains of susceptible mice (129 and C57BL/6). We then compared the gene expression patterns in these mice with 129 mice carrying a null mutation in the epithelial-restricted integrin beta6 subunit (beta6(-/-)), which develop inflammation but are protected from pulmonary fibrosis. Cluster analysis identified two distinct groups of genes involved in the inflammatory and fibrotic responses. Analysis of gene expression at multiple time points after bleomycin administration revealed sequential induction of subsets of genes that characterize each response. The availability of this comprehensive data set should accelerate the development of more effective strategies for intervention at the various stages in the development of fibrotic diseases of the lungs and other organs.
Figures
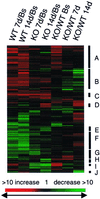
Figure 1
Cluster analysis of 470 genes that changed >2-fold and had at least one mean intensity value of 100 in at least one of the experimental conditions. The data are presented as a ratio of the mean of the average intensities from all animals in each experimental group. Genes that were present at higher levels in the examined group (increased) are shown in progressively brighter shades of red, depending on the fold difference, and genes that were expressed at lower levels are shown in progressively brighter shades of green. Genes shown in black were not different between the two groups being compared. Columns identify the pair-wise comparison being made between wild-type (WT) or _β6_−/− (KO) mice examined at baseline (Bs) or at 7 days (7d) or 14 days (14d) after treatment with intratracheal bleomycin. Baseline values included both untreated mice (n = 4) and mice sacrificed 7 days after treatment with intratracheal saline (n = 3). All other values were the mean of three average intensities. Cluster analysis was performed on log transformed values of the fold ratios with
gene cluster
and presented with
treeview
. The letters on the right define distinct clusters that are discussed in the text or presented in the following figures or on our web site.
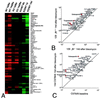
Figure 2
(A) Enlargement of Cluster B from Fig. 1 of genes that were not different at baseline and were induced preferentially in wild-type mice. The name of each gene is shown to the right of each row. Genes shown in bold text were induced more than 8-fold in response to bleomycin. (B) Because many of the genes in cluster B had been previously shown to be induced by TGF-β, expression of 53 known TGF-β-inducible genes was compared 14 days after bleomycin treatment in _β6_−/− mice (x axis) and wild-type (β6+/+) mice (y axis). Genes induced at least 8-fold by bleomycin are shown bold in these panels, and the mean intensity values are shown by red boxes. The middle diagonal in B and C is the line of identity. (C) Comparison of expression of the same 53 TGF-β-inducible genes at baseline (x axis) and 14 days after bleomycin (y axis) in wild-type C57BL/6 mice.
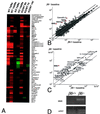
Figure 3
(A) Enlargement of Cluster A from Fig. 1. The name of each gene is shown to the right of each row. Genes shown in bold text were, at baseline, more than 8-fold higher in _β_6−/− mice than in _β_6+/+ mice. (B) Scatter plot comparing the values of all the genes in _β_6+/+ (x axis) and _β_6−/− mice (y axis). Genes shown in bold were expressed at least 8-fold higher in _β_6−/− mice compared with _β_6+/+ mice, and their mean intensity values are shown by red circles. The middle diagonal in B and C is the line of identity. The upper and lower diagonals represent 2-fold increases and decreases, respectively. (C) Scatter plot comparing the expression of 47 genes restricted to leukocytes in _β_6+/+ mice and _β_6−/− mice. (D) Ethidium bromide stained 2.5% agarose gel demonstrating the results of noncompetitive RT-PCR with primers specific for MME from macrophages purified by adherence from bronchoalveolar lavage of _β_6+/+ mice or _β_6−/− mice. (Bottom) Parallel competitive RT-PCR with primers and a plasmid competitor specific for HPRT.
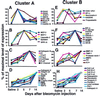
Figure 4
Time course of expression after saline and at days 2, 5, 7, and 14 after bleomycin, in C57BL/6 mice of selected genes from Clusters A (A–D) and B (E and F) that were also induced by bleomycin in this strain. Each panel shows data for genes that followed a similar temporal pattern of induction. Values for each gene are plotted as a percentage of maximal induction for that gene, which is considered 100%.
Similar articles
- Common and distinct mechanisms of induced pulmonary fibrosis by particulate and soluble chemical fibrogenic agents.
Dong J, Yu X, Porter DW, Battelli LA, Kashon ML, Ma Q. Dong J, et al. Arch Toxicol. 2016 Feb;90(2):385-402. doi: 10.1007/s00204-015-1589-3. Epub 2015 Sep 7. Arch Toxicol. 2016. PMID: 26345256 Free PMC article. - Integrin-mediated activation of transforming growth factor-beta(1) in pulmonary fibrosis.
Sheppard D. Sheppard D. Chest. 2001 Jul;120(1 Suppl):49S-53S. doi: 10.1378/chest.120.1_suppl.s49. Chest. 2001. PMID: 11451914 - Molecular monitoring of bleomycin-induced pulmonary fibrosis by cDNA microarray-based gene expression profiling.
Katsuma S, Nishi K, Tanigawara K, Ikawa H, Shiojima S, Takagaki K, Kaminishi Y, Suzuki Y, Hirasawa A, Ohgi T, Yano J, Murakami Y, Tsujimoto G. Katsuma S, et al. Biochem Biophys Res Commun. 2001 Nov 9;288(4):747-51. doi: 10.1006/bbrc.2001.5853. Biochem Biophys Res Commun. 2001. PMID: 11688970 - The transcription factor NRF2 protects against pulmonary fibrosis.
Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. Cho HY, et al. FASEB J. 2004 Aug;18(11):1258-60. doi: 10.1096/fj.03-1127fje. Epub 2004 Jun 18. FASEB J. 2004. PMID: 15208274 - Effects of montelukast, a cysteinyl-leukotriene type 1 receptor antagonist, on the pathogenesis of bleomycin-induced pulmonary fibrosis in mice.
Shimbori C, Shiota N, Okunishi H. Shimbori C, et al. Eur J Pharmacol. 2011 Jan 10;650(1):424-30. doi: 10.1016/j.ejphar.2010.09.084. Epub 2010 Oct 27. Eur J Pharmacol. 2011. PMID: 21034736
Cited by
- Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis.
Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Dakhlallah D, et al. Am J Respir Crit Care Med. 2013 Feb 15;187(4):397-405. doi: 10.1164/rccm.201205-0888OC. Epub 2013 Jan 10. Am J Respir Crit Care Med. 2013. PMID: 23306545 Free PMC article. - Antifibrotic effect of disulfiram on bleomycin-induced lung fibrosis in mice and its impact on macrophage infiltration.
Okabe Y, Toda E, Urushiyama H, Terashima Y, Kunugi S, Kajimoto Y, Terasaki M, Matsushima K, Saito A, Yamauchi Y, Nagase T, Shimizu A, Terasaki Y. Okabe Y, et al. Sci Rep. 2024 Oct 10;14(1):23653. doi: 10.1038/s41598-024-71770-z. Sci Rep. 2024. PMID: 39384840 Free PMC article. - Mesenchymal stem cell engraftment in lung is enhanced in response to bleomycin exposure and ameliorates its fibrotic effects.
Ortiz LA, Gambelli F, McBride C, Gaupp D, Baddoo M, Kaminski N, Phinney DG. Ortiz LA, et al. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8407-11. doi: 10.1073/pnas.1432929100. Epub 2003 Jun 18. Proc Natl Acad Sci U S A. 2003. PMID: 12815096 Free PMC article. - Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling.
Zhao X, Nicholls JM, Chen YG. Zhao X, et al. J Biol Chem. 2008 Feb 8;283(6):3272-3280. doi: 10.1074/jbc.M708033200. Epub 2007 Nov 30. J Biol Chem. 2008. PMID: 18055455 Free PMC article. - Intravenous immunoglobulin and fibrosis.
Molina V, Blank M, Shoenfeld Y. Molina V, et al. Clin Rev Allergy Immunol. 2005 Dec;29(3):321-6. doi: 10.1385/CRIAI:29:3:321. Clin Rev Allergy Immunol. 2005. PMID: 16391408 Review.
References
- Coultas D B, Zumwalt R E, Black W C, Sobonya R E. Am J Respir Crit Care Med. 1994;150:967–972. - PubMed
- Ryu J H, Colby T V, Hartman T E. Mayo Clin Proc. 1998;73:1085–1101. - PubMed
- King T E., Jr Ann Intern Med. 1998;129:806–812. - PubMed
- Snider G L, Hayes J A, Korthy A L. Am Rev Respir Dis. 1978;117:1099–1108. - PubMed
- Shen A S, Haslett C, Feldsien D C, Henson P M, Cherniack R M. Am Rev Respir Dis. 1988;137:564–571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL33259/HL/NHLBI NIH HHS/United States
- HL47412/HL/NHLBI NIH HHS/United States
- R37 HL053949/HL/NHLBI NIH HHS/United States
- F32 HL009793/HL/NHLBI NIH HHS/United States
- R01 HL053949/HL/NHLBI NIH HHS/United States
- AI33259/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases