Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli - PubMed (original) (raw)
Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli
K Amor et al. Infect Immun. 2000 Mar.
Abstract
In the lipopolysaccharides of Escherichia coli there are five distinct core oligosaccharide (core OS) structures, designated K-12 and R1 to R4. The objective of this work was to determine the prevalences of these core OS types within the species. Unique sequences in the waa (core OS biosynthesis) gene operon were used to develop a PCR-based system that facilitated unequivocal determination of the core OS types in isolates of E. coli. This system was applied to the 72 isolates in the E. coli ECOR collection, a compilation of isolates that is considered to be broadly representative of the genetic diversity of the species. Fifty (69. 4%) of the ECOR isolates contained the R1 core OS, 8 (11.1%) were representatives of R2, 8 (11.1%) were R3, 2 (2.8%) were R4, and only 4 (5.6%) were K-12. R1 is the only core OS type found in all four major phylogenetic groups (A, B1, B2, and D) in the ECOR collection. Virulent extraintestinal pathogenic E. coli isolates tend to be closely related to group B2 and, to a lesser extent, group D isolates. All of the ECOR representatives from the B2 and D groups had the R1 core OS. In contrast, commensal E. coli isolates are more closely related to group A, which contains isolates representing each of the five core OS structures. R3 was the only core OS type found in 38 verotoxigenic E. coli (VTEC) isolates from humans and cattle belonging to the common enterohemorrhagic E. coli serogroups O157, O111, and O26. Although isolates from other VTEC serogroups showed more core OS diversity, the R3 type (83.1% of all VTEC isolates) was still predominant. When non-VTEC commensal isolates from cattle were analyzed, it was found that most possessed the R1 core OS type.
Figures
FIG. 1
Structures of the five known outer core OSs from the LPSs of E. coli and genetic organization of the central waaQ operon from each of the waa (core OS biosynthesis) loci. HepII is the last residue of the inner core OS. The asterisks indicate the points of attachment of O antigen, but this has only been determined experimentally for the R1 and R2 core OSs. The gene products responsible for each residue of the core OS are indicated. Details can be found elsewhere (16). PCR products diagnostic for each core OS type are shown below the relevant physical maps. The primer sequences are given in Table 1.
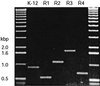
FIG. 2
Agarose gel showing the electrophoretic mobilities of PCR products obtained from the prototype strains representing each core OS type.
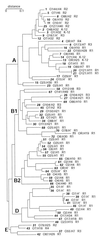
FIG. 3
Phylogenetic tree of the ECOR isolates (17) showing the distribution of the five core OS types. The number of the ECOR isolate is given in boldface italics. For each isolate the determined O:H serotype and core OS type are listed. OR, R-LPS; ?, antigen not determined. The serotyping results reported here showed some differences to those reported elsewhere (T. S. Whittam,
http: //www.bio.psu.edu/People/Faculty/Whittam/Lab
). In many cases the discrepancies reflect a nontypeable reaction in one set of data or the other and may reflect subtle differences in growth conditions, protocol, or antisera. In the analysis reported here, there were significantly less nontypeable antigens. In some cases, the serotypes varied between the two studies. To verify the data reported here, two independently held ECOR collections were examined and each of the isolates giving different serotypes was retested.
Similar articles
- Distribution of lipopolysaccharide core types among avian pathogenic Escherichia coli in relation to the major phylogenetic groups.
Dissanayake DR, Wijewardana TG, Gunawardena GA, Poxton IR. Dissanayake DR, et al. Vet Microbiol. 2008 Dec 10;132(3-4):355-63. doi: 10.1016/j.vetmic.2008.05.024. Epub 2008 Jul 2. Vet Microbiol. 2008. PMID: 18597955 - Virulence and antimicrobial resistance determinants of verotoxigenic Escherichia coli (VTEC) and of multidrug-resistant E. coli from foods of animal origin illegally imported to the EU by flight passengers.
Nagy B, Szmolka A, Smole Možina S, Kovač J, Strauss A, Schlager S, Beutlich J, Appel B, Lušicky M, Aprikian P, Pászti J, Tóth I, Kugler R, Wagner M. Nagy B, et al. Int J Food Microbiol. 2015 Sep 16;209:52-9. doi: 10.1016/j.ijfoodmicro.2015.06.026. Epub 2015 Jul 3. Int J Food Microbiol. 2015. PMID: 26148965 - The expression of an R3 lipopolysaccharide-core by pathotypes of Escherichia coli.
Chart H, Perry NT, Jenkins C. Chart H, et al. J Appl Microbiol. 2004;96(5):982-6. doi: 10.1111/j.1365-2672.2004.02233.x. J Appl Microbiol. 2004. PMID: 15078514 - Role of non-O157 VTEC.
Bettelheim KA. Bettelheim KA. Symp Ser Soc Appl Microbiol. 2000;(29):38S-50S. doi: 10.1111/j.1365-2672.2000.tb05331.x. Symp Ser Soc Appl Microbiol. 2000. PMID: 10880178 Review. - Molecular insights into the assembly and diversity of the outer core oligosaccharide in lipopolysaccharides from Escherichia coli and Salmonella.
Whitfield C, Kaniuk N, Frirdich E. Whitfield C, et al. J Endotoxin Res. 2003;9(4):244-9. doi: 10.1179/096805103225001440. J Endotoxin Res. 2003. PMID: 12935355 Review.
Cited by
- Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates.
Han W, Wu B, Li L, Zhao G, Woodward R, Pettit N, Cai L, Thon V, Wang PG. Han W, et al. J Biol Chem. 2012 Feb 17;287(8):5357-65. doi: 10.1074/jbc.M111.308486. Epub 2011 Dec 12. J Biol Chem. 2012. PMID: 22158874 Free PMC article. - Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1.
Šimoliūnas E, Vilkaitytė M, Kaliniene L, Zajančkauskaitė A, Kaupinis A, Staniulis J, Valius M, Meškys R, Truncaitė L. Šimoliūnas E, et al. Viruses. 2015 Nov 27;7(12):6163-81. doi: 10.3390/v7122932. Viruses. 2015. PMID: 26633460 Free PMC article. - A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917.
Grozdanov L, Zähringer U, Blum-Oehler G, Brade L, Henne A, Knirel YA, Schombel U, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Rietschel ET, Dobrindt U. Grozdanov L, et al. J Bacteriol. 2002 Nov;184(21):5912-25. doi: 10.1128/JB.184.21.5912-5925.2002. J Bacteriol. 2002. PMID: 12374825 Free PMC article. - Lipopolysaccharide endotoxins.
Raetz CR, Whitfield C. Raetz CR, et al. Annu Rev Biochem. 2002;71:635-700. doi: 10.1146/annurev.biochem.71.110601.135414. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045108 Free PMC article. Review. - PCR for specific detection of H7 flagellar variant of fliC among extraintestinal pathogenic Escherichia coli.
Johnson JR, Stell AL. Johnson JR, et al. J Clin Microbiol. 2001 Oct;39(10):3712-7. doi: 10.1128/JCM.39.10.3712-3717.2001. J Clin Microbiol. 2001. PMID: 11574599 Free PMC article.
References
- Aleksic S. WHO report on Shiga-like toxin producing Escherichia coli (SLTEC), with emphasis on zoonotic aspects. Geneva, Switzerland: World Health Organization; 1995.
- Appelmelk B J, An Y-Q, Hekker T A M, Thijs L G, MacLaren D M, deGraaf J. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology. 1994;140:1119–1124. - PubMed
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources