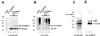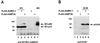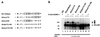Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b - PubMed (original) (raw)
Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b
H Hofmann et al. J Virol. 2000 Mar.
Abstract
The 86-kDa IE2 protein (IE2-p86) of human cytomegalovirus (HCMV) is a potent transactivator of viral as well as cellular promoters. Several lines of evidence indicate that this broad transactivation spectrum is mediated by protein-protein interactions. To identify novel cellular binding partners, we performed a yeast two-hybrid screen using a N-terminal deletion mutant of IE2-p86 comprising amino acids 135 to 579 as a bait. Here, we report the isolation of two ubiquitin-homologous proteins, SUMO-1 and hSMT3b, as well as their conjugating activity hUBC9 (human ubiquitin-conjugating enzyme 9) as specific interaction partners of HCMV IE2. The polypeptides SUMO-1 and hSMT3b have previously been shown to be covalently coupled to a subset of nuclear proteins such as the nuclear domain 10 (ND10) proteins PML and Sp100 in a manner analogous to ubiquitinylation, which we call SUMOylation. By Western blot analysis, we were able to show that the IE2-p86 protein can be partially converted to a 105-kDa isoform in a dose-dependent manner after cotransfection of an epitope-tagged SUMO-1. Immunoprecipitation experiments of the conjugated isoforms using denaturing conditions further confirmed the covalent coupling of SUMO-1 or hSMT3b to IE2-p86 both after transient transfection and after lytic infection of human primary fibroblasts. Moreover, we defined two modification sites within IE2, located in an immediate vicinity at amino acid positions 175 and 180, which appear to be used alternatively for coupling. By using a SUMOylation-defective mutant, we showed that the targeting of IE2-p86 to ND10 occurs independent of this modification. However, a strong reduction of IE2-mediated transactivation of two viral early promoters and a heterologous promoter was observed in cotransfection analysis with the SUMOylation-defective mutant. This suggests a functional relevance of covalent modification by ubiquitin-homologous proteins for IE2-mediated transactivation, possibly by providing an additional interaction motif for cellular cofactors.
Figures
FIG. 1
Specific interaction between HCMV IE2(135-579) and hUBC9, hSMT3b, and SUMO-1 in yeast. Yeast cells were transformed with two separate vectors, one of which encoded either IE2(135-579) fused to the GAL4 DNA-binding domain (IE2pAS1) or the DNA-binding domain alone (pAS1). The second plasmid encoded either the GAL4 activation domain alone (pACT) or hUBC9, hSMT3b, and SUMO-1 as fusions with the GAL4 activation domain, (plasmids hUBC9 pACT, hSMT3b pACT, and SUMO-1 pACT, respectively). Yeast colonies were selected for the presence of both plasmids with dropout media lacking tryptophan and leucine and subsequently analyzed for the expression of β-galactosidase. The association of murine p53 (encoded by plasmid pVA3; Clontech) and SV40 large T antigen (plasmid pTD1; Clontech) served as a positive control. (A) Qualitative analysis of interactions between IE2 (135-579) and hUBC9, hSMT3b, and SUMO-1 as determined in filter lift experiments after staining for β-galactosidase activity. (B) Quantitation of the association between IE2(135-579) and hUBC9, hSMT3b, and SUMO-1 as determined by liquid β-galactosidase assays (ONPG assays). Cotransformation experiments were performed as for panel A. β-Galactosidase activity was assayed from liquid cultures in at least three independent experiments, each with duplicate samples. The β-galactosidase activity of yeast cells transformed with a vector combination encoding murine p53 and SV40 large T antigen was set as 100.
FIG. 2
Covalent interaction between IE2-p86 and FLAG-SUMO-1 in 293 cells. 293 cells were transfected either with the empty eucaryotic expression vector pCB6, the FLAG-SUMO-1 expression vector, or plasmid pHM134 (encoding IE2-p86) either alone or in combination as indicated. Cells in panels A and B were lysed using NP-40 lysis buffer, whereas lysates in panels C and D were prepared under denaturing conditions using SDS lysis buffer. The lysates were fractionated by SDS-PAGE (8% gel) and analyzed by Western blotting. (A) Western blot analysis of cell lysates using the IE2-p86 polyclonal antiserum. (B) Incubation of the same filter membrane as in panel A, using the anti-FLAG MAb. Lanes: 1, transfection with expression vector pCB6 alone; 2, transfection with the FLAG-SUMO-1 expression vector alone; 3, transfection with plasmid pHM134 (encoding IE2-p86) alone; 4 to 6, transfection with a constant amount of plasmid pHM134 and increasing amounts of plasmid pFLAG-SUMO-1. (C and D) Western blot analyses of cell lysates using the IE2-p86 polyclonal antiserum (C) or MAb 810 (D). Lanes: 1, transfection with vector pCB6 alone; 2, transfection with plasmid pHM134 alone; 3, transfection with a combination of plasmids pHM134 and pFLAG-SUMO-1. The IE2-p86 modification by an endogenous UbH moiety is indicated by a closed circle; the modification by cotransfected FLAG-SUMO-1 is depicted by an open circle.
FIG. 3
Analysis of covalent modification of HCMV IE1-p72 and pUL84 by FLAG-SUMO-1 and FLAG-hSMT3b, respectively. 293 cells were transfected with expression vectors encoding IE1-p72, pUL84, or FLAG-SUMO-1/hSMT3b as indicated and lysed in SDS lysis buffer. Cell extracts were resolved by SDS-PAGE (10% gel) and subjected to immunoblotting using MAb 810 directed against a N-terminal epitope shared by IE1-p72 and IE2-p86 (A) or a polyclonal antiserum directed against HCMV pUL84 (B). (A) Lanes: 1, transfection with the IE1-p72 expression vector alone; 2, transfection with a combination of vectors encoding IE1-p72 and FLAG-SUMO-1; 3, transfection with a combination of vectors encoding IE1-p72 and FLAG-hSMT3b; 4, transfection with vector pCB6 alone; 5, transfection with plasmid pFLAG-SUMO-1 alone; 6, transfection with plasmid pFLAG-hSMT3b alone; 7, transfection with plasmid pHM134 (encoding IE2-p86) alone. (B) Lanes: 1, transfection with vector pCB6 alone; 2, transfection with plasmid pFLAG-SUMO-1 alone; 3, transfection with plasmid pFLAG-hSMT3b alone; 4, transfection with UL84 expression vector pcDNAUL84 alone; 5, transfection with a combination of pcDNAUL84 and pFLAG-SUMO-1; 6, transfection with a combination of pcDNAUL84 and pFLAG-hSMT3b. Sizes are indicated in kilodaltons.
FIG. 4
Evidence for a covalent modification of HCMV IE2-p86 by SUMO-1 and hSMT3b in immunoprecipitation experiments. 293 cells were transfected with an expression vector encoding IE2-p86, FLAG-SUMO-1/hSMT3b, or FLAG-UL84 as indicated and prepared for immunoprecipitation as described in Materials and Methods. Immunoprecipitations were performed with the IE2-p86 antiserum or the IE2-p86 preimmune serum, as indicated by bars. Precipitates were washed three times and separated by SDS-PAGE (10% gel). Thereafter, coprecipitated interactor proteins were detected by Western blot analysis using the anti-FLAG MAb. The interaction between IE2-p86 and FLAG-UL84 served as a positive control. Asterisks depict the two additional anti-FLAG reactive bands in IE2-UbH precipitates. (A) Western blot analysis of precipitated IE2-hSMT3b conjugates. Lanes: 1, cell lysis under native lysis conditions; 2 to 8, cell lysis under denaturating conditions. Transfection was performed with plasmids encoding FLAG-UL84 and IE2-p86 (lanes 1 and 2), IE2-p86 alone (lane 3), FLAG-hSMT3b alone (lane 4), and IE2-p86 and FLAG-hSMT3b (lanes 5 to 8). (B) Western blot analysis of precipitated IE2-SUMO-1 conjugates. Lanes: 1, cell lysis under native lysis conditions; 2 to 5, cell lysis under denaturating conditions. Transfection was performed with plasmids encoding FLAG-UL84 and IE2-p86 (lane 1), FLAG-SUMO-1 alone (lane 2), FLAG-SUMO-1 and IE2-p86 (lanes 3 and 4), and IE2-p86 alone (lane 5). Here and in subsequent figures, sizes are indicated in kilodaltons, and IgG and IP stand for immunoglobulin G and immunoprecipitation, respectively.
FIG. 5
Evidence for a covalent modification of HCMV IE2-p86 in infected HFFs. (A) Western blot analysis using lysates from infected HFFs and transfected 293 cells. HFFs were either mock infected or infected with the laboratory strain AD169 for the indicated time periods. Cells were lysed in SDS sample buffer, and the lysates were fractionated by SDS-PAGE (10% gel) followed by Western blot analysis using the anti-pHM178 polyclonal antiserum directed against exon 5 of IE2-p86. Lanes: 1, lysate from mock-infected HFFs; 2 to 6, lysates from HFFs that were infected with HCMV for 6, 24, 30, 48, and 72 h, respectively; 7 to 8; lysates from HFFs that were infected with HCMV for 48 and 72 h, respectively; 9, lysate from 293 cells that were transfected with the IE2-p86 expression vector pHM134. The 105-kDa isoform of IE2 is indicated by a closed circle. (B to D) Immunoprecipitation of IE1 or IE2 conjugates from HCMV-infected HFFs. HFFs were transfected with an expression vector encoding FLAG-SUMO-1 or FLAG-hSMT3b. The day after transfection, cells were either mock infected or infected with HCMV strain AD169 for 72 h as indicated. Thereafter, immunoprecipitation was performed with MAb 810, recognizing an epitope common to both IE2-p86 and IE1-p72. Western blot analysis was performed using the anti-FLAG MAb in order to detect SUMO-1 or hSMT3b (B), the IE2-p86 polyclonal antiserum in order to detect IE2 proteins (C), and MAb p63-27 in order to detect IE1 proteins (D). Transfection of HFFs was performed with plasmids encoding either FLAG-SUMO-1 (lanes 1 and 2) or FLAG-hSMT3b (lane 3). The modification of IE2-p86 by the FLAG-UbH proteins is indicated by open circles.
FIG. 6
Mapping of lysine residues responsible for SUMOylation of HCMV IE2-p86. (A) Schematic overview depicting the IE2 deletion mutants used. Lysine residues in IE2-p86 are indicated by arrows; the positions of nuclear localization signals (NLS) within IE2 are also indicated. The eucaryotic expression vectors were transfected into 293 cells either alone or in combination with FLAG-SUMO-1 and analyzed for conjugation by Western blotting, indicated by + (conjugation detectable) or − (no conjugation observed). The IE2 mutants comprising amino acids 290 to 548, 310 to 548, and 329 to 548 were cloned into the pSuperCATCH-NLS vector to ensure nuclear localization. (B) Sequence similarity analysis between already determined SUMOylation sites in cellular proteins and HCMV IE2-p86. The proposed SUMOylation consensus motif is indicated by boxes; the lysine residues responsible for isopeptide bond formation are shaded darker.
FIG. 7
Interaction of IE2-p86 amino acid mutants with UbH proteins. (A) Schematic overview of the amino acid deletions or substitutions in IE2-p86 generated by PCR mutagenesis. Potential SUMOylation consensus motifs are indicated by boxes. (B) Western blot analysis of the individual IE2-p86 mutants. 293 cells were transfected with expression vectors encoding either wild-type IE2-p86 or the individual mutants alone or in combination with a vector encoding FLAG-SUMO-1 as indicated. Cell lysates were analyzed by immunoblotting using the IE2-p86 polyclonal antiserum. Lanes: 1, transfection with a vector encoding FLAG-SUMO-1 alone; 2, transfection with IE2-p86 expression vector pHM134; 3, transfection with pHM134 and pFLAG-SUMO-1; 4, transfection with a vector encoding IE2mut175 alone; 5, transfection with pIE2mut175 and pFLAG-SUMO-1; 6, transfection with a vector encoding IE2mut180 alone; 7, transfection with pIE2mut180 and pFLAG-SUMO-1; 8, transfection with a vector encoding IE2del174-181 alone; 9, transfection with pIE2del174-181 and pFLAG-SUMO-1; 10, transfection with a vector encoding IE2mut175+180 alone; 11, transfection with pIE2mut175+180 and pFLAG-SUMO-1. The IE2-p86 modification by an endogenous UbH moiety is indicated by a closed circle; the modification by cotransfected FLAG-SUMO-1 is depicted by an open circle.
FIG. 8
Analysis of IE2-p86 mutants by immunoprecipitation experiments. 293 cells were transfected with the indicated expression vectors, and immunoprecipitations were performed using the IE2-p86 polyclonal antiserum or preimmune serum (A) or MAb 810 (panel B). (A) Conjugation of IE2-p86 mutants with UbH proteins. Coprecipitated interactor proteins were detected in Western blot experiments using the anti-FLAG MAb M2. For lanes 5, 9, 13, and 17, cell lysis was performed with NP-40 lysis buffer; for all other lanes, SDS-containing buffer was used (see Materials and Methods). (B) Evidence for dimerization capability of the individual IE2-p86 mutants. 293 cells were transfected with a vector encoding an N-terminally truncated IE2-protein [FLAG-IE2(135-579)] in combination with expression vectors for either IE2-p86 wild-type protein (wtIE2) or a mutated variant of IE2 (IE2mut175, IE2mut180, IE2del174-181, or IE2mut175+180). Cells were lysed in NP-40 lysis buffer, and immunoprecipitations were performed with MAb 810, which recognizes an N-terminal epitope within IE2-p86 or the individual mutants that are not contained in the FLAG-IE2(135-579) protein (see panel C). Thereafter, Western blot analysis was performed using the anti-FLAG MAb in order to specifically detect the FLAG-IE2(135-579) protein.
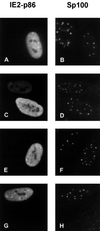
FIG. 9
Subcellular localization of IE2-p86 and mutant IE2 proteins in HFFs. HFFs grown on coverslips were transfected with expression vectors encoding the wild-type IE2-p86 protein fused to GFP (A and B) or IE2 mutant IE2mut175 (C and D), IE2mut180 (E and F), or IE2mut175+180 (G and H). Indirect immunofluorescence was performed with MAb 810 and an FITC-conjugated anti-mouse secondary antibody in order to detect the IE2 mutants (C, E, and G). Wild-type IE2 was detected through its GFP moiety (A). Additionally, costaining was performed with a polyclonal Sp26 antiserum directed against Sp100 and a TRITC-conjugated anti-rabbit conjugated secondary antibody in order to detect ND10 (B, D, F, and H).
FIG. 10
Luciferase analysis after cotransfection of luciferase reporter constructs carrying viral early promoters or the HIV-1 LTR with expression plasmids for various IE2 mutants. U373MG cells were transfected with luciferase reporter constructs carrying the viral early promoters of the UL112/113 gene region (A, lanes 1 to 7), the UL84 gene (B, lanes 1 to 6), or the HIV LTR (C, lanes 1 to 6). Lanes: 1, cotransfection was performed with the empty expression vector pCB6; 2, cotransfection was performed with expression vector pHM134 encoding wild-type IE2-p86; 3, cotransfection was performed with the expression vector for IE2 mutant IE2mut175; 4, cotransfection was performed with the expression vector for IE2 mutant IE2mut180; 5, cotransfection was performed with the expression vector for IE2 mutant IE2mut175+180; 6, cotransfection was performed with the internal deletion mutant IE2del174-181; 7, cotransfection was performed with the internal deletion mutant IE86ΔSX (kindly provided by D. Spector) (56). Each experiment was performed in triplicate and was repeated at least three times. Fold activation was calculated relative to the basal activity of each reporter construct after cotransfection with the empty pCB6 vector. (D) Western blot analysis of 293 cell extracts after transfection of various IE2 expression plasmids using the IE2-specific MAb 810. Lanes: 1, transfection was performed with expression vector pCB6; 2, transfection was performed with vector pHM134; 3, transfection was performed with the vector encoding IE2mut175; 4, transfection was performed with the vector encoding IE2mut180; 5, transfection was performed with the vector encoding IE2mut175+180; 6, transfection was performed with the internal deletion mutant IE2del174-181; 7, transfection was performed with the internal deletion mutant IE86ΔSX.
Similar articles
- Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9.
Ahn JH, Xu Y, Jang WJ, Matunis MJ, Hayward GS. Ahn JH, et al. J Virol. 2001 Apr;75(8):3859-72. doi: 10.1128/JVI.75.8.3859-3872.2001. J Virol. 2001. PMID: 11264375 Free PMC article. - The ND10 Component Promyelocytic Leukemia Protein Acts as an E3 Ligase for SUMOylation of the Major Immediate Early Protein IE1 of Human Cytomegalovirus.
Reuter N, Schilling EM, Scherer M, Müller R, Stamminger T. Reuter N, et al. J Virol. 2017 Apr 28;91(10):e02335-16. doi: 10.1128/JVI.02335-16. Print 2017 May 15. J Virol. 2017. PMID: 28250117 Free PMC article. - The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication.
Gebert S, Schmolke S, Sorg G, Flöss S, Plachter B, Stamminger T. Gebert S, et al. J Virol. 1997 Sep;71(9):7048-60. doi: 10.1128/JVI.71.9.7048-7060.1997. J Virol. 1997. PMID: 9261435 Free PMC article. - Intracellular targeting of proteins by sumoylation.
Wilson VG, Rangasamy D. Wilson VG, et al. Exp Cell Res. 2001 Nov 15;271(1):57-65. doi: 10.1006/excr.2001.5366. Exp Cell Res. 2001. PMID: 11697882 Review. - New insights into the role of the small ubiquitin-like modifier (SUMO) in plants.
Park HJ, Yun DJ. Park HJ, et al. Int Rev Cell Mol Biol. 2013;300:161-209. doi: 10.1016/B978-0-12-405210-9.00005-9. Int Rev Cell Mol Biol. 2013. PMID: 23273862 Review.
Cited by
- IE1 of Human Cytomegalovirus Inhibits Necroptotic Cell Death via Direct and Indirect Modulation of the Necrosome Complex.
Heusel AT, Rapp S, Stamminger T, Scherer M. Heusel AT, et al. Viruses. 2024 Feb 13;16(2):290. doi: 10.3390/v16020290. Viruses. 2024. PMID: 38400065 Free PMC article. - Post-Translational Modifications of Kaposi's Sarcoma-Associated Herpesvirus Regulatory Proteins - SUMO and KSHV.
Campbell M, Izumiya Y. Campbell M, et al. Front Microbiol. 2012 Feb 14;3:31. doi: 10.3389/fmicb.2012.00031. eCollection 2012. Front Microbiol. 2012. PMID: 22347876 Free PMC article. - Characterization of a human cytomegalovirus with phosphorylation site mutations in the immediate-early 2 protein.
Heider JA, Yu Y, Shenk T, Alwine JC. Heider JA, et al. J Virol. 2002 Jan;76(2):928-32. doi: 10.1128/jvi.76.2.928-932.2002. J Virol. 2002. PMID: 11752183 Free PMC article. - Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover.
Hardeland U, Steinacher R, Jiricny J, Schär P. Hardeland U, et al. EMBO J. 2002 Mar 15;21(6):1456-64. doi: 10.1093/emboj/21.6.1456. EMBO J. 2002. PMID: 11889051 Free PMC article. - Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5.
Sun D, Xu P, He B. Sun D, et al. J Virol. 2011 Oct;85(19):10261-8. doi: 10.1128/JVI.00389-11. Epub 2011 Jul 27. J Virol. 2011. PMID: 21795356 Free PMC article.
References
- Alford C. Breast milk transmission of cytomegalovirus (CMV) infection. Adv Exp Med Biol. 1991;310:293–299. - PubMed
- Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1981–2010.
- Andreoni M, Faircloth M, Vugler L, Britt W J. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods. 1989;23:157–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous