ATP-dependent chromatin-remodeling complexes - PubMed (original) (raw)
Review
ATP-dependent chromatin-remodeling complexes
M Vignali et al. Mol Cell Biol. 2000 Mar.
No abstract available
Figures
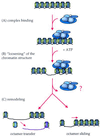
FIG. 1
Two-step model of SWI/SNF and RSC action in chromatin remodeling. The binding of the remodeling complex to chromatin is ATP independent (A). Upon ATP addition, the conformation of nucleosomes changes as a consequence of the alteration of histone-DNA interactions (B). This disruption results in remodeling of the chromatin (C), which might occur while the complex is still bound or might persist after it is released from the chromatin (indicated by the question mark). Remodeling that occurs may result in transfer of histone octamers to different DNA segments in trans or in sliding of the octamers in cis (i.e., to a different position in the same DNA molecule). The exact consequence of remodeling is likely dependent on the exact context of nucleosomes at a given promoter and can lead to either (i) activation of transcription or (ii) repression.
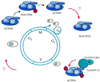
FIG. 2
Cell cycle regulation of the SWI/SNF complex. The activity of hSWI/SNF is regulated, at least in part, by phosphorylation of some of its subunits. The complex is activated after G1 by a cyclin E/cdk2-dependent phosphorylation event of BAF155 and BRG1. Phosphorylation toward the end of G2, which might also occur at the level of BRG1, inactivates it, while a dephosphorylation even occurring late in G2 also seems to have an activating role in SWI/SNF function. The question marks denote the lack of information concerning how these two apparently contradictory sets of data are related.
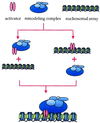
FIG. 3
Targeting of the SWI/SNF complex. The remodeling complexes are directed to sites in chromatin via their interactions with transcriptional regulatory factors. The available data do not distinguish between the two models presented. It is possible that the factor binds the DNA first and then acts as a docking pad for the remodeling complex. Alternatively, it is possible that the interaction between the factor and the remodeling complex takes place in solution and the DNA-binding domain of the transcription factor directs the remodeling complex to chromatin in a later step.
Similar articles
- Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling.
Wang GG, Allis CD, Chi P. Wang GG, et al. Trends Mol Med. 2007 Sep;13(9):373-80. doi: 10.1016/j.molmed.2007.07.004. Epub 2007 Sep 5. Trends Mol Med. 2007. PMID: 17822959 Free PMC article. Review. - Purification and characterization of a human factor that assembles and remodels chromatin.
LeRoy G, Loyola A, Lane WS, Reinberg D. LeRoy G, et al. J Biol Chem. 2000 May 19;275(20):14787-90. doi: 10.1074/jbc.C000093200. J Biol Chem. 2000. PMID: 10747848 - ATP-dependent chromatin remodeling enzymes and their various roles in cell cycle control.
Morettini S, Podhraski V, Lusser A. Morettini S, et al. Front Biosci. 2008 May 1;13:5522-32. doi: 10.2741/3096. Front Biosci. 2008. PMID: 18508602 Review. - ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo.
Aoyagi S, Trotter KW, Archer TK. Aoyagi S, et al. Vitam Horm. 2005;70:281-307. doi: 10.1016/S0083-6729(05)70009-1. Vitam Horm. 2005. PMID: 15727808 Review. - Cajal body dynamics and association with chromatin are ATP-dependent.
Platani M, Goldberg I, Lamond AI, Swedlow JR. Platani M, et al. Nat Cell Biol. 2002 Jul;4(7):502-8. doi: 10.1038/ncb809. Nat Cell Biol. 2002. PMID: 12068306
Cited by
- The BAF complex enhances transcription through interaction with H3K56ac in the histone globular domain.
Hyun K, Ahn J, Kim H, Kim J, Kim YI, Park HS, Roeder RG, Lee JE, Kim J. Hyun K, et al. Nat Commun. 2024 Nov 7;15(1):9614. doi: 10.1038/s41467-024-53981-0. Nat Commun. 2024. PMID: 39511190 Free PMC article. - Mechanisms of DNA Damage Response in Mammalian Oocytes.
Sun F, Sutovsky P, Patterson AL, Balboula AZ. Sun F, et al. Adv Anat Embryol Cell Biol. 2024;238:47-68. doi: 10.1007/978-3-031-55163-5_3. Adv Anat Embryol Cell Biol. 2024. PMID: 39030354 Review. - Acute depletion of BRG1 reveals its primary function as an activator of transcription.
Ren G, Ku WL, Ge G, Hoffman JA, Kang JY, Tang Q, Cui K, He Y, Guan Y, Gao B, Liu C, Archer TK, Zhao K. Ren G, et al. Nat Commun. 2024 May 29;15(1):4561. doi: 10.1038/s41467-024-48911-z. Nat Commun. 2024. PMID: 38811575 Free PMC article. - Epigenetic Regulation in Schizophrenia: Focus on Methylation and Histone Modifications in Human Studies.
Delphin N, Aust C, Griffiths L, Fernandez F. Delphin N, et al. Genes (Basel). 2024 Feb 21;15(3):272. doi: 10.3390/genes15030272. Genes (Basel). 2024. PMID: 38540331 Free PMC article. Review. - Genome organization across scales: mechanistic insights from in vitro reconstitution studies.
Oberbeckmann E, Oudelaar AM. Oberbeckmann E, et al. Biochem Soc Trans. 2024 Apr 24;52(2):793-802. doi: 10.1042/BST20230883. Biochem Soc Trans. 2024. PMID: 38451192 Free PMC article. Review.
References
- Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor NCoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. - PubMed
- Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. - PubMed
- Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. - PubMed
- Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources