Physical association of ubiquitin ligases and the 26S proteasome - PubMed (original) (raw)
Physical association of ubiquitin ligases and the 26S proteasome
Y Xie et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The ubiquitin (Ub) system recognizes degradation signals of the target proteins through the E3 components of E3-E2 Ub ligases. A targeted substrate bears a covalently linked multi-Ub chain and is degraded by the ATP-dependent 26S proteasome, which consists of the 20S core protease and two 19S particles. The latter mediate the binding and unfolding of a substrate protein before its transfer to the interior of the 20S core. It is unclear how a targeted substrate is delivered to the 26S proteasome, inasmuch as Rpn10p, the only known proteasomal subunit that binds multi-Ub chains, has been found to be not essential for degradation of many proteins in the yeast Saccharomyces cerevisiae. Here we show that Ubr1p and Ufd4p, the E3 components of two distinct Ub ligases, directly interact with the 26S proteasome. Specifically, Ubr1p is shown to bind to the Rpn2p, Rpt1p, and Rpt6p proteins of the 19S particle, and Ufd4p is shown to bind to Rpt6p. These and related results suggest that a substrate-bound Ub ligase participates in the delivery of substrates to the proteasome, because of affinity between the ligase's E3 component and specific proteins of the 19S particle.
Figures
Figure 1
Toxicity of overexpressed N-end rule pathway is decreased by overexpressed Rpn1p and enhanced by overexpressed Rpt1p or Rpt6p. The_S. cerevisiae_ strain JD52 (23) was transformed with pKM1313, a low-copy, URA3_-based plasmid expressing_UBR1 and RAD6 from the bidirectional PGAL1,10 promoter (24). The pKM1313-containing JD52 cells were transformed either with the pRS425 vector (A),RPN1 (expressed from its own promoter in pRS425) (B), RPT1 (expressed from the PCUP1 promoter in p314CUP1RPT1) (C), or_RPT6_ (expressed from the PCUP1 promoter in p314CUP1RPT6) (D). (E and_F_) JD52 cells carrying both pKM1313 and the_RPN1_-expressing plasmid were further transformed with p314CUP1RPT1 (E) and p314CUP1RPT6 (F), respectively. Shown here are plates with cells grown in the galactose-containing minimal medium containing 0.2 mM CuSO4 for 4 days at 30°C. On dextrose-containing plates (no overexpression of Ubr1p and Rad6p), all transformants grew at similar rates, irrespective of the presence of overexpressed Rpn1p, Rpt1p, or Rpt6p (data not shown).
Figure 2
Overexpression of Rpn1p inhibits degradation of N-end rule substrates. A UPR-based pulse–chase assay was used (see Results).S. cerevisiae JD52 (UBR1) cells coexpressing either DHFR-HA-UbR48-Leu-βgal (A) or DHFR-HA-UbR48-His-βgal (B), and either Rpn1p (expressed from its own promoter in pRS425) or pRS425 alone were labeled with [35S]methionine for 5 min at 30°C, followed by a chase for 0, 10, and 30 min. Cell extracts were immunoprecipitated with both anti-βgal and anti-HA antibodies, followed by SDS/12% PAGE, autoradiography, and quantitation using PhosphorImager (Molecular Dynamics). (C) His-βgal and Leu-βgal decay curves calculated from the UPR-based data in A and_B_ as described (28, 32). The bands of X-βgals and the reference protein DHFR-HA-UbR48 (“ref”) are indicated.
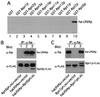
Figure 3
Ubr1p, the E3 of the N-end rule pathway, is physically associated with the 26S proteasome. (A and B) Ubr1p interacts with Rpn2p, Rpt1p, and Rpt6p in GST-pulldown assays. Extracts of S. cerevisiae containing overexpressed FLAG-Ubr1p (A) or the purified FLAG-Ubr1p protein (B) were incubated with glutathione-agarose beads preloaded with the indicated GST fusions. The retained proteins were eluted, fractionated by SDS/8% PAGE, and immunoblotted with anti-FLAG antibody. Approximately equal amounts of different GST fusions were immobilized on glutathione-agarose beads in these assays, as verified by Coomassie staining (data not shown). (C and D) In vivo association of Ubr1p and Rpt6p. Extracts of S. cerevisiae AVY107 (_ubr1_Δ) expressing either both FLAG-Ubr1p and HA-Rpt6p, or FLAG-Ubr1p alone, or HA-Rpt6p alone were incubated with anti-FLAG antibody (C) or anti-HA antibody (D). The immunoprecipitated proteins were separated by SDS/12% PAGE and transferred to nitrocellulose membrane. The top halves of C and D show the results of immunoblotting with anti-FLAG antibody; the bottom halves show the analogous data with anti-HA antibody. (E) Coimmunoprecipitation of Pre6p and Ubr1p. Extracts of S. cerevisiae AVY107 (_ubr1_Δ) expressing both FLAG-Ubr1p and HA-Pre6p, FLAG-Ubr1p alone, or HA-Pre6p alone were incubated with anti-HA antibody, followed by the immunoprecipitation/immunoblotting described in C and D.
Figure 4
Rpn1p interacts with Rpt1p, Rpt6p, and Rpn10p. _E. coli_-expressed Rpn1p-FLAG (A) and Rpn10p-FLAG (B), both carrying the FLAG epitope at the C terminus, were incubated with glutathione-agarose beads preloaded with the indicated GST fusions. The retained proteins were fractionated by SDS/PAGE, followed by immunoblotting with anti-FLAG antibody. Approximately equal amounts of different GST fusions were immobilized on glutathione-agarose beads in these assays, as verified by Coomassie staining (data not shown).
Figure 5
Ufd4p, the E3 of the UFD pathway, is physically associated with the proteasome. (A) Ufd4p interacts with Rpt6p in the GST-pulldown assay. Extracts of S. cerevisiae containing overexpressed HA-Ufd4p were incubated with glutathione-agarose beads preloaded with different GST fusions, as indicated. The retained proteins were eluted, fractionated by SDS/8% PAGE, and immunoblotted with anti-HA antibody. Approximately equal amounts of different GST fusions were immobilized on glutathione-agarose beads in these assays, as verified by Coomassie staining (data not shown). (B) In vivo association of Ufd4p and Rpt6p. Extracts of S. cerevisiae JD52 (UBR1) expressing both HA-Ufd4p and Rpt6p-FLAG, HA-Ufd4p alone, or Rpt6p-FLAG alone were incubated with anti-HA antibody. The immunoprecipitated proteins were separated by SDS/10% PAGE and transferred to nitrocellulose membrane. (Upper) The results of immunoblotting with anti-HA antibody. (Lower) The data with anti-FLAG antibody. (C) Coimmunoprecipitation of Rpn1p and Ufd4p. Extracts of S. cerevisiae JD52 expressing both HA-Ufd4p and Rpn1p-FLAG, HA-Ufd4p alone, or Rpn1p-FLAG alone were incubated with anti-HA antibody, followed by the immunoprecipitation/immunoblotting procedure described in B, except that SDS/7% PAGE was used.
Similar articles
- UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis.
Xie Y, Varshavsky A. Xie Y, et al. Nat Cell Biol. 2002 Dec;4(12):1003-7. doi: 10.1038/ncb889. Nat Cell Biol. 2002. PMID: 12447385 - Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes.
Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Verma R, et al. Mol Biol Cell. 2000 Oct;11(10):3425-39. doi: 10.1091/mbc.11.10.3425. Mol Biol Cell. 2000. PMID: 11029046 Free PMC article. - Ubiquitin biology: an old dog learns an old trick.
Pickart CM. Pickart CM. Nat Cell Biol. 2000 Aug;2(8):E139-41. doi: 10.1038/35019610. Nat Cell Biol. 2000. PMID: 10934481 - Roles of ubiquitin-mediated proteolysis in cell cycle control.
Hershko A. Hershko A. Curr Opin Cell Biol. 1997 Dec;9(6):788-99. doi: 10.1016/s0955-0674(97)80079-8. Curr Opin Cell Biol. 1997. PMID: 9425343 Review. - The 26S proteasome: ubiquitin-mediated proteolysis in the tunnel.
Kierszenbaum AL. Kierszenbaum AL. Mol Reprod Dev. 2000 Oct;57(2):109-10. doi: 10.1002/1098-2795(200010)57:2<109::AID-MRD1>3.0.CO;2-9. Mol Reprod Dev. 2000. PMID: 10984410 Review.
Cited by
- The deubiquitinase Rpn11 functions as an allosteric ubiquitin sensor to promote substrate engagement by the 26S proteasome.
Htet ZM, Dong KC, Martin A. Htet ZM, et al. bioRxiv [Preprint]. 2024 Oct 24:2024.10.24.620116. doi: 10.1101/2024.10.24.620116. bioRxiv. 2024. PMID: 39484543 Free PMC article. Preprint. - BIG enhances Arg/N-degron pathway-mediated protein degradation to regulate Arabidopsis hypoxia responses and suberin deposition.
Zhang H, Rundle C, Winter N, Miricescu A, Mooney BC, Bachmair A, Graciet E, Theodoulou FL. Zhang H, et al. Plant Cell. 2024 Sep 3;36(9):3177-3200. doi: 10.1093/plcell/koae117. Plant Cell. 2024. PMID: 38608155 Free PMC article. - Analysis of Proteasome-Associated Ubiquitin Ligase Activity.
Wang Z, Orosa-Puente B, Spoel SH. Wang Z, et al. Methods Mol Biol. 2023;2581:57-67. doi: 10.1007/978-1-0716-2784-6_5. Methods Mol Biol. 2023. PMID: 36413310 - Development and progression of cancer cachexia: Perspectives from bench to bedside.
Lim S, Brown JL, Washington TA, Greene NP. Lim S, et al. Sports Med Health Sci. 2020 Dec;2(4):177-185. doi: 10.1016/j.smhs.2020.10.003. Epub 2020 Dec 3. Sports Med Health Sci. 2020. PMID: 34447946 Free PMC article. - Structural basis for the N-degron specificity of ClpS1 from Arabidopsis thaliana.
Kim L, Heo J, Kwon DH, Shin JS, Jang SH, Park ZY, Song HK. Kim L, et al. Protein Sci. 2021 Mar;30(3):700-708. doi: 10.1002/pro.4018. Epub 2020 Dec 30. Protein Sci. 2021. PMID: 33368743 Free PMC article.
References
- Maniatis T. Genes Dev. 1999;13:505–510. - PubMed
- Nasmyth K. Trends Biochem Sci. 1999;24:98–104. - PubMed
- Hershko A, Ciechanover A. Annu Rev Biochem. 1998;76:425–479. - PubMed
- Peters J-M, King R W, Deshaies R J. In: Ubiquitin and the Biology of the Cell. Peters J-M, Harris J R, Finley D, editors. New York: Plenum; 1998. pp. 345–387.
- Varshavsky A. Trends Biochem Sci. 1997;22:383–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 DK039520/DK/NIDDK NIH HHS/United States
- R01 GM031530/GM/NIGMS NIH HHS/United States
- R01 DK039520/DK/NIDDK NIH HHS/United States
- DK39520/DK/NIDDK NIH HHS/United States
- R37 DK039520/DK/NIDDK NIH HHS/United States
- GM31530/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases