Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif - PubMed (original) (raw)
. 2000 Feb 15;14(4):435-51.
Affiliations
- PMID: 10691736
- PMCID: PMC316385
Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif
S Germain et al. Genes Dev. 2000.
Abstract
We have investigated the regulation of the activin-inducible distal element (DE) of the Xenopus goosecoid promoter. The results show that paired-like homeodomain transcription factors of the Mix family, Mixer and Milk, but not Mix.1, mediate activin/TGF-beta-induced transcription through the DE by interacting with the effector domain of Smad2, thereby recruiting active Smad2/Smad4 complexes to the Mixer/Milk-binding site. We identify a short motif in the carboxyl termini of Mixer and Milk, which is demonstrated to be both necessary and sufficient for interaction with the effector domain of Smad2 and is required for mediating activin/TGF-beta-induced transcription. This motif is not confined to these homeodomain proteins, but is also present in the Smad2-interacting winged-helix proteins Xenopus Fast-1, human Fast-1, and mouse Fast-2. We demonstrate directly that transcription factors of different DNA-binding specificity recruit activated Smads to distinct promoter elements via a common mechanism. These observations, together with the temporal and spatial expression patterns of Mixer and Milk, lead us to propose a model for mesoendoderm formation in Xenopus in which these homeodomain transcription factor/Smad complexes play a role in initiating and maintaining transcription of target genes in response to endogenous activin-like signals.
Figures
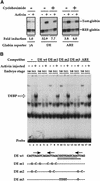
Figure 1
Activin-responsive transcription via the goosecoid DE. (A) Activin-responsive transcription through the DE is partially dependent on new protein synthesis. One-cell embryos were injected with REF–globin internal control together with globin reporters driven by the minimal γ-actin promoter (γA), or by multiple copies of the DE or ARE upstream of the mimimal promoter. Animal caps, cut at stage 8 were cultured for 6 hr with or without activin in the absence or presence of 5 μg/ml cycloheximide. Globin transcripts from reporter genes (Test-globin) or the internal control (REF-globin) were detected by RNase protection (Howell and Hill 1997). Transcriptional activation was calculated as a ratio of the levels of Test-globin to REF-globin. Activin-induced transcription is expressed as fold inductions. (B) An activin-inducible protein (DEBP) binds the goosecoid DE. Whole-cell extracts prepared from stage 8 or stage 11 embryos or stage 11 embryos overexpressing activin, were analyzed by bandshift assay using the single DE probe. Activin-inducible protein DEBP is indicated. Competitor oligonucleotides were used at a 50-fold molar excess over probe where indicated. (Bottom) Sequences of wild-type DE and mutant oligonucleotides indicating only the altered nucleotides. The paired-like homeodomain-binding site comprising two inverted TAAT motifs (Wilson et al. 1993) is denoted by arrows; a third homeodomain core-binding site at the 3′ end is also indicated by an arrow. (Thick dotted line) Sequence reminiscent of a half-site for T-box protein (AGGTGTGAAATT) (Kispert et al. 1995); (underline) almost perfect binding site for the ZFH-1 family of zinc finger homeodomain proteins (AGGTGAGCAA) (Funahashi et al. 1993). (C) Formation of DEBP requires new protein synthesis. Extracts were made from uninjected stage 8 embryos (lane 1), stage 10.5 embryos (lanes 2,3), or stage 10.5 embryos overexpressing activin (lanes 4,5) and analyzed by bandshift using the DE probe. Where indicated, embryos had been preincubated in cycloheximide before stage 8.
Figure 1
Activin-responsive transcription via the goosecoid DE. (A) Activin-responsive transcription through the DE is partially dependent on new protein synthesis. One-cell embryos were injected with REF–globin internal control together with globin reporters driven by the minimal γ-actin promoter (γA), or by multiple copies of the DE or ARE upstream of the mimimal promoter. Animal caps, cut at stage 8 were cultured for 6 hr with or without activin in the absence or presence of 5 μg/ml cycloheximide. Globin transcripts from reporter genes (Test-globin) or the internal control (REF-globin) were detected by RNase protection (Howell and Hill 1997). Transcriptional activation was calculated as a ratio of the levels of Test-globin to REF-globin. Activin-induced transcription is expressed as fold inductions. (B) An activin-inducible protein (DEBP) binds the goosecoid DE. Whole-cell extracts prepared from stage 8 or stage 11 embryos or stage 11 embryos overexpressing activin, were analyzed by bandshift assay using the single DE probe. Activin-inducible protein DEBP is indicated. Competitor oligonucleotides were used at a 50-fold molar excess over probe where indicated. (Bottom) Sequences of wild-type DE and mutant oligonucleotides indicating only the altered nucleotides. The paired-like homeodomain-binding site comprising two inverted TAAT motifs (Wilson et al. 1993) is denoted by arrows; a third homeodomain core-binding site at the 3′ end is also indicated by an arrow. (Thick dotted line) Sequence reminiscent of a half-site for T-box protein (AGGTGTGAAATT) (Kispert et al. 1995); (underline) almost perfect binding site for the ZFH-1 family of zinc finger homeodomain proteins (AGGTGAGCAA) (Funahashi et al. 1993). (C) Formation of DEBP requires new protein synthesis. Extracts were made from uninjected stage 8 embryos (lane 1), stage 10.5 embryos (lanes 2,3), or stage 10.5 embryos overexpressing activin (lanes 4,5) and analyzed by bandshift using the DE probe. Where indicated, embryos had been preincubated in cycloheximide before stage 8.
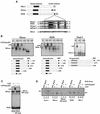
Figure 2
DEBP specifically interacts with the effector domain of Smad2. (A) Whole-cell extracts prepared from either stage 8 embryos (lanes 1–4), stage 10.5 embryos (lanes 5–8), or stage 10.5 embryos overexpressing activin (lanes 9–12) were analyzed by bandshift using the DE probe. Extracts were mixed with either purified GST protein (100 ng) (lanes 4,8,12) or two concentrations (20 and 100 ng) of purified GSTSmad2C (lanes 2, 3, 6, 7, 10, 11) before addition of probe. (Open arrow) DEBP; (black arrow) GSTSmad2C bound to DEBP. (B) The interaction of DEBP with Smad2C is specific. The assay was as in A using GST fusions of Smad1C, Smad2C, or Smad4C as indicated.
Figure 2
DEBP specifically interacts with the effector domain of Smad2. (A) Whole-cell extracts prepared from either stage 8 embryos (lanes 1–4), stage 10.5 embryos (lanes 5–8), or stage 10.5 embryos overexpressing activin (lanes 9–12) were analyzed by bandshift using the DE probe. Extracts were mixed with either purified GST protein (100 ng) (lanes 4,8,12) or two concentrations (20 and 100 ng) of purified GSTSmad2C (lanes 2, 3, 6, 7, 10, 11) before addition of probe. (Open arrow) DEBP; (black arrow) GSTSmad2C bound to DEBP. (B) The interaction of DEBP with Smad2C is specific. The assay was as in A using GST fusions of Smad1C, Smad2C, or Smad4C as indicated.
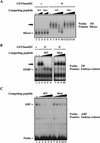
Figure 3
Homeodomain proteins, Mixer and Milk, but not Mix.1, interact with Smad2C. (A) Mixer and Milk are good candidates for the endogenous protein DEBP. Whole-cell extracts were prepared from stage 10.5 embryos or embryos injected at the 1-cell stage with mRNA encoding myc-tagged Mixer, Milk, Mix.1, or activin, and DE-binding activity was assayed by bandshift. Anti-myc antibody or purified GSTSmad2C were added where indicated. (Open arrow) DEBP; (gray arrow) supershifted complexes. (B) Interaction of GSTSmad2C with members of the Mix family and Fast-1. In vitro-translated Mixer, Milk, Mix.1, and Fast.1 were assayed by bandshift for their interaction with purified GSTSmad2C or GST using the appropriate radiolabeled DE or ARE probes. Transcription factors complexed with probe are indicated; (black arrow) ternary complex with GSTSmad2C.
Figure 4
Characterization of the SIM. (A) Schematics of Mix.1, Mixer, and Milk, with the conserved homeodomains and a carboxy-terminal acidic domain indicated. (Black box) A region conserved in Milk and Mixer, also present in Xenopus Fast-1 and mouse Fast-2 (expanded below where the black line denotes the boundaries of the conserved sequences). (Black shading) Identical amino acids; (gray shading) similar amino acids. The numbers indicate the positions of these amino acids in the full-length sequences of the individual proteins. (B) Carboxy-terminal deletion mutants of Mixer, Milk, and Fast-1 (schematized below) were produced in vitro and their interaction with GSTSmad2C assayed by bandshift using the DE or ARE probe as appropriate. Complexes of transcription factors and probe are indicated; (black arrow) ternary complex with GSTSmad2C. Smad interaction motif (SIM) and DNA-binding domains are indicated. Note that Milk gives rise to two complexes, both of which shift with GSTSmad2C, which correspond to a dimer of Milk and a higher order complex. (C) Mutation of the prolines in the PPNK core motif abolishes the interaction with Smad2C. Full-length Mixer or a mutant derivative (Mixer PP mut), in which the two prolines in the PPNK-containing motif are mutated to alanines, were produced in vitro and assayed for interaction with GSTSmad2C by bandshift using the DE probe. (D) Interaction of Mixer, Milk, and Fast-1 with Smad2C in solution. 35S-labeled transcription factors as indicated were incubated with Sepharose-bound GST (lanes 2,5,8,11,14) or GSTSmad2C (lanes 3,6,9,12,15) and bound protein was visualized by SDS-PAGE and autoradiography. A fraction of input protein was analyzed for comparison (lanes 1,4,7,10,13).
Figure 5
The Smad interaction motif is sufficient to interact with Smad2. (A) A peptide containing the SIM of Mixer competes specifically for interaction of Mixer with Smad2C. In vitro-translated Mixer was incubated with DE probe alone (lane 1) or in the presence of 1 or 10 pmoles of wild-type peptide (lanes 2,3) or mutant peptide (lanes 4,5). GSTSmad2C (20 ng) was included in the reactions in lanes 6–14, with the addition of 0.3, 1, 3, or 10 pmoles of wild-type peptide (lanes 7–10) or mutant peptide (lanes 11–14). Mixer complexed with probe is indicated; (black arrow), ternary complex with GSTSmad2C. (B) A peptide containing the Mixer SIM specifically disrupts the interaction of DEBP with Smad2C. Whole-cell extracts made from activin-injected stage 10.5 embryos were analyzed by bandshift assay with the DE probe in the absence (lane 1) or presence of GSTSmad2C (20 ng; lanes 2–10) with the addition of 5, 15, 30, and 100 pmoles wild-type peptide (lanes 3–6) or mutant peptide (lanes 7–10). DEBP is indicated; (black arrow) ternary complex with GSTSmad2C. (C) A peptide containing the Mixer SIM specifically disrupts the formation of ARF. Whole-cell extracts made from activin-injected stage 8 embryos were analyzed by bandshift assay with the ARE probe in the absence (lane 1) or presence of 10, 30, 60, 100, or 200 pmoles of wild-type peptide (lanes 2–6) or mutant peptide (lanes 7–11). The endogenous ARF complex is indicated.
Figure 6
Mixer and Milk interact with activated Smads in vivo. (A) Mixer forms a ligand-dependent complex with Smad2 and Smad4 in solution. Extracts were prepared from NIH-3T3 cells transfected with myc–Smad2, myc–Smad4, and either Flag–Fast-1, Flag–Mixer, or a Flag-tagged mutant derivative (Mixer PP mut), which had been incubated with or without TGF-β1 (2 ng/ml) for 1 hr. Extracts were assayed either by immunoprecipitation of complexes with anti-Flag antibody followed by Western blotting with anti-Myc antibody (top), or Western blotting the whole extract with anti-Flag antibody (middle), or with anti-Myc antibody (bottom). (B,C) Fast-1 and Mixer form ligand-dependent complexes on DNA with endogenous Smad2 and Smad4. Extracts were prepared from NIH-3T3 cells transfected with Flag-tagged Fast-1, Mixer, or Mixer (PP mut), which had been incubated with or without TGF-β1 (2 ng/ml) for 1 hr. Extracts were analyzed by bandshift assay on the ARE (B) or DE (C) probe. Anti-flag, anti-Smad2, or anti-Smad4 antibodies were included in the binding reactions where indicated. In B ARF and antibody-supershifted ARF are indicated. In C, Mixer or Mixer (PP mut) bound to probe, the Mixer–Smad complex and antibody-supershifted Mixer–Smad complex are indicated.
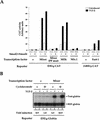
Figure 7
Mixer and Milk mediate TGF-β-dependent transcriptional activation through the DE. (A) NIH-3T3 cells were transfected with the CAT reporters, and plasmids expressing transcription factors, Smad2 and Smad4, as indicated. Cells were cultured with or without TGF-β1 (2 ng/ml) for 8 hr. Cells were harvested and CAT activity measured relative to lacZ activity from the internal control. The data are from a representative experiment, and similar results were obtained in other independent experiments. (B) Mixer mediates TGF-β-dependent transcriptional activation through the DE in the absence of protein synthesis. NIH-3T3 cells were transfected with the (DE)4-globin reporter and REF-globin internal control with or without Mixer expression plasmid. Cells were cultured with or without TGF-β1 (2 ng/ml) for 4 hr in the absence or presence of 50 μg/ml cycloheximide. Globin transcripts from the reporter genes (Test-globin) or the internal control (REF-globin) were detected by RNase protection and quantitated as in Fig. 1.
Figure 8
The temporal and spatial expression patterns of Mixer and Milk in Xenopus embryos makes them good candidates for mediating transcription of goosecoid in response to an endogenous activin-like signal. (A) Coexpression of goosecoid with Mixer and Milk at early gastrula stages. Xenopus embryos were fixed at stage 10.25, cut in half through the middle of the dorsal lip, and processed for in situ hybridization with probes against goosecoid, Mixer, Milk, or Cerberus (as a control to visualize the prospective mesoendodermal boundary). (Arrowhead) Blastopore lip. The mRNAs are visualized with a dark blue/purple stain. It is clear that goosecoid is expressed on the dorsal side in prospective mesodermal and endodermal cells expressing Mixer and/or Milk. (Right) Schematic of stage 10.25 embryo cut in the same way as the embryo halves. (Red) Prospective notochord; (orange) prospective head mesoderm; (pink) somitic mesoderm; (light orange) ventral migrating mesoderm; (yellow) prospective endoderm (adapted from Keller 1991). (B) Time course of expression of goosecoid (Gsc), Mixer, Milk, and the FGF receptor (FGFR) assayed by RNase protection. The Milk probe also detects Bix3 (Tada et al. 1998). Embryos were sampled at stage 8 and subsequent times indicated. In lanes 9–16 the embryos had been preincubated with cycloheximide from 30 min before stage 8. (C) A model showing that TGF-β/activin-activated Smads translocate to the nucleus, where they interact with homeodomain transcription factors, Mixer and Milk through the SIM to activate transcription. (D) A model indicating the proposed role of the Mixer/Milk/Bix3-Smad complexes in the formation of mesoderm and endoderm in early Xenopus embryos. (Black arrows) Induction of gene expression; (gray arrows) activation of protein complexes. Milk/Bix3–Smad complexes are involved in the initiation of transcription of mesoendodermal genes and Mixer/Milk/Bix–Smad complexes are involved in the maintenance of gene expression. For discussion, see text.
Figure 8
The temporal and spatial expression patterns of Mixer and Milk in Xenopus embryos makes them good candidates for mediating transcription of goosecoid in response to an endogenous activin-like signal. (A) Coexpression of goosecoid with Mixer and Milk at early gastrula stages. Xenopus embryos were fixed at stage 10.25, cut in half through the middle of the dorsal lip, and processed for in situ hybridization with probes against goosecoid, Mixer, Milk, or Cerberus (as a control to visualize the prospective mesoendodermal boundary). (Arrowhead) Blastopore lip. The mRNAs are visualized with a dark blue/purple stain. It is clear that goosecoid is expressed on the dorsal side in prospective mesodermal and endodermal cells expressing Mixer and/or Milk. (Right) Schematic of stage 10.25 embryo cut in the same way as the embryo halves. (Red) Prospective notochord; (orange) prospective head mesoderm; (pink) somitic mesoderm; (light orange) ventral migrating mesoderm; (yellow) prospective endoderm (adapted from Keller 1991). (B) Time course of expression of goosecoid (Gsc), Mixer, Milk, and the FGF receptor (FGFR) assayed by RNase protection. The Milk probe also detects Bix3 (Tada et al. 1998). Embryos were sampled at stage 8 and subsequent times indicated. In lanes 9–16 the embryos had been preincubated with cycloheximide from 30 min before stage 8. (C) A model showing that TGF-β/activin-activated Smads translocate to the nucleus, where they interact with homeodomain transcription factors, Mixer and Milk through the SIM to activate transcription. (D) A model indicating the proposed role of the Mixer/Milk/Bix3-Smad complexes in the formation of mesoderm and endoderm in early Xenopus embryos. (Black arrows) Induction of gene expression; (gray arrows) activation of protein complexes. Milk/Bix3–Smad complexes are involved in the initiation of transcription of mesoendodermal genes and Mixer/Milk/Bix–Smad complexes are involved in the maintenance of gene expression. For discussion, see text.
Figure 8
The temporal and spatial expression patterns of Mixer and Milk in Xenopus embryos makes them good candidates for mediating transcription of goosecoid in response to an endogenous activin-like signal. (A) Coexpression of goosecoid with Mixer and Milk at early gastrula stages. Xenopus embryos were fixed at stage 10.25, cut in half through the middle of the dorsal lip, and processed for in situ hybridization with probes against goosecoid, Mixer, Milk, or Cerberus (as a control to visualize the prospective mesoendodermal boundary). (Arrowhead) Blastopore lip. The mRNAs are visualized with a dark blue/purple stain. It is clear that goosecoid is expressed on the dorsal side in prospective mesodermal and endodermal cells expressing Mixer and/or Milk. (Right) Schematic of stage 10.25 embryo cut in the same way as the embryo halves. (Red) Prospective notochord; (orange) prospective head mesoderm; (pink) somitic mesoderm; (light orange) ventral migrating mesoderm; (yellow) prospective endoderm (adapted from Keller 1991). (B) Time course of expression of goosecoid (Gsc), Mixer, Milk, and the FGF receptor (FGFR) assayed by RNase protection. The Milk probe also detects Bix3 (Tada et al. 1998). Embryos were sampled at stage 8 and subsequent times indicated. In lanes 9–16 the embryos had been preincubated with cycloheximide from 30 min before stage 8. (C) A model showing that TGF-β/activin-activated Smads translocate to the nucleus, where they interact with homeodomain transcription factors, Mixer and Milk through the SIM to activate transcription. (D) A model indicating the proposed role of the Mixer/Milk/Bix3-Smad complexes in the formation of mesoderm and endoderm in early Xenopus embryos. (Black arrows) Induction of gene expression; (gray arrows) activation of protein complexes. Milk/Bix3–Smad complexes are involved in the initiation of transcription of mesoendodermal genes and Mixer/Milk/Bix–Smad complexes are involved in the maintenance of gene expression. For discussion, see text.
Similar articles
- Xenopus Smad4beta is the co-Smad component of developmentally regulated transcription factor complexes responsible for induction of early mesodermal genes.
Howell M, Itoh F, Pierreux CE, Valgeirsdottir S, Itoh S, ten Dijke P, Hill CS. Howell M, et al. Dev Biol. 1999 Oct 15;214(2):354-69. doi: 10.1006/dbio.1999.9430. Dev Biol. 1999. PMID: 10525340 - Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2.
Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Labbé E, et al. Mol Cell. 1998 Jul;2(1):109-20. doi: 10.1016/s1097-2765(00)80119-7. Mol Cell. 1998. PMID: 9702197 - The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis.
Yeo CY, Chen X, Whitman M. Yeo CY, et al. J Biol Chem. 1999 Sep 10;274(37):26584-90. doi: 10.1074/jbc.274.37.26584. J Biol Chem. 1999. PMID: 10473623 - SIP1 (Smad interacting protein 1) and deltaEF1 (delta-crystallin enhancer binding factor) are structurally similar transcriptional repressors.
van Grunsven LA, Schellens A, Huylebroeck D, Verschueren K. van Grunsven LA, et al. J Bone Joint Surg Am. 2001;83-A Suppl 1(Pt 1):S40-7. J Bone Joint Surg Am. 2001. PMID: 11263664 Review. - The transcriptional role of Smads and FAST (FoxH1) in TGFbeta and activin signalling.
Attisano L, Silvestri C, Izzi L, Labbé E. Attisano L, et al. Mol Cell Endocrinol. 2001 Jun 30;180(1-2):3-11. doi: 10.1016/s0303-7207(01)00524-x. Mol Cell Endocrinol. 2001. PMID: 11451566 Review.
Cited by
- Transcriptional integration of Wnt and Nodal pathways in establishment of the Spemann organizer.
Reid CD, Zhang Y, Sheets MD, Kessler DS. Reid CD, et al. Dev Biol. 2012 Aug 15;368(2):231-41. doi: 10.1016/j.ydbio.2012.05.018. Epub 2012 May 22. Dev Biol. 2012. PMID: 22627292 Free PMC article. - Xenopus as a model system to study transcriptional regulatory networks.
Koide T, Hayata T, Cho KW. Koide T, et al. Proc Natl Acad Sci U S A. 2005 Apr 5;102(14):4943-8. doi: 10.1073/pnas.0408125102. Epub 2005 Mar 28. Proc Natl Acad Sci U S A. 2005. PMID: 15795378 Free PMC article. Review. - Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation.
Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Levy L, et al. Mol Cell Biol. 2007 Sep;27(17):6068-83. doi: 10.1128/MCB.00664-07. Epub 2007 Jun 25. Mol Cell Biol. 2007. PMID: 17591695 Free PMC article. - In control of biology: of mice, men and Foxes.
Wijchers PJ, Burbach JP, Smidt MP. Wijchers PJ, et al. Biochem J. 2006 Jul 15;397(2):233-46. doi: 10.1042/BJ20060387. Biochem J. 2006. PMID: 16792526 Free PMC article. Review. - A comparative analysis of Smad-responsive motifs identifies multiple regulatory inputs for TGF-β transcriptional activation.
Itoh Y, Koinuma D, Omata C, Ogami T, Motizuki M, Yaguchi SI, Itoh T, Miyake K, Tsutsumi S, Aburatani H, Saitoh M, Miyazono K, Miyazawa K. Itoh Y, et al. J Biol Chem. 2019 Oct 18;294(42):15466-15479. doi: 10.1074/jbc.RA119.009877. Epub 2019 Sep 3. J Biol Chem. 2019. PMID: 31481467 Free PMC article.
References
- Blumberg B, Wright CV, De Robertis EM, Cho KW. Organizer-specific homeobox genes in Xenopus laevis embryos. Science. 1991;253:194–196. - PubMed
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382:595–601. - PubMed
- Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-β signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. - PubMed
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. - PubMed
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous