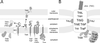Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems - PubMed (original) (raw)
Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems
A M Grahn et al. J Bacteriol. 2000 Mar.
Abstract
During bacterial conjugation, the single-stranded DNA molecule is transferred through the cell envelopes of the donor and the recipient cell. A membrane-spanning transfer apparatus encoded by conjugative plasmids has been proposed to facilitate protein and DNA transport. For the IncPalpha plasmid RP4, a thorough sequence analysis of the gene products of the transfer regions Tra1 and Tra2 revealed typical features of mainly inner membrane proteins. We localized essential RP4 transfer functions to Escherichia coli cell fractions by immunological detection with specific polyclonal antisera. Each of the gene products of the RP4 mating pair formation (Mpf) system, specified by the Tra2 core region and by traF of the Tra1 region, was found in the outer membrane fraction with one exception, the TrbB protein, which behaved like a soluble protein. The membrane preparation from Mpf-containing cells had an additional membrane fraction whose density was intermediate between those of the cytoplasmic and outer membranes, suggesting the presence of attachment zones between the two E. coli membranes. The Tra1 region is known to encode the components of the RP4 relaxosome. Several gene products of this transfer region, including the relaxase TraI, were detected in the soluble fraction, but also in the inner membrane fraction. This indicates that the nucleoprotein complex is associated with and/or assembled facing the cytoplasmic site of the E. coli cell envelope. The Tra1 protein TraG was predominantly localized to the cytoplasmic membrane, supporting its potential role as an interface between the RP4 Mpf system and the relaxosome.
Figures
FIG. 1
Schematic representation of potential membrane-spanning or membrane-associating segments within RP4 transfer proteins. Polypeptide chains of proteins encoded by RP4 transfer regions are shown as lines corresponding to the scale at the top of the figure. The number of amino acid (AA) residues deduced from the nucleotide sequence (44) is given for each polypeptide. Signal sequences were predicted according to the method of von Heijne (64) and are shown as dark-gray bars. Arrows indicate the predicted cleavage sites. Potential transmembrane helices (white bars) were found by the PHDhtm and the PHDtopology programs (European Molecular Biology Laboratory [EMBL], Heidelberg, Germany [47, 48]). Additional stretches of at least 10 consecutive hydrophobic amino acid residues (horizontally striped bars) were identified by the hydropathy plot method of Kyte and Doolittle (33). Potential amphiphilic helical segments (vertically striped bars) were determined by using the secondary-structure prediction program PHDsec (EMBL) (–51) in combination with the helical-wheel projection method of Schiffer and Edmundson (54). A surplus of positively charged amino acid residues, which is important for the topology of membrane proteins according to the positive-inside rule (65), is indicated by a plus sign.
FIG. 2
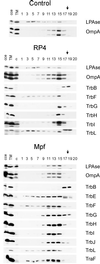
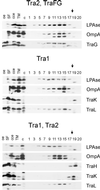
Immunological detection of RP4 transfer proteins in E. coli cell fractions. SCS1 cells containing the appropriate plasmids were fractionated into soluble (SF), CM, and OM fractions as described in Materials and Methods. Plasmids in the host strain encoding the indicated transfer functions or individual gene products are listed in Table 2. Proteins in the soluble and the membrane fractions were separated on standard SDS-polyacrylamide gels and were transferred to PVDF membranes, which were subsequently incubated with the appropriate antiserum specific for Tra2 (A) and Tra1 (B) region gene products (Table 3). To the lanes on the left side of each panel were applied appropriate amounts of crude cell extract (cce) from the respective expression strains.
FIG. 3
Immunological detection of RP4 Mpf components in the membrane fractions. Flotation of the total membranes in density gradients resulted in the separation of the CM and the OM of the control SCS1 strain. An additional, an intermediate light-scattering zone (MCM) was detected in cells containing either RP4 or the Mpf genes (plasmids pML123 and pWP471). Aliquots of the fractions were analyzed in standard SDS-polyacrylamide gels, and the proteins were transferred to a PVDF membrane. Incubation with specific antisera identified the Mpf components (listed on the right) in different cell fractions. cce, crude cell extract; TM, total membranes; c, crude cell extract of the control SCS1 strain; 1 to 20, fractions collected starting from the top of the gradient. The arrows indicate the sample application position.
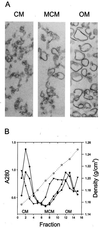
FIG. 4
(A) Thin-section electron microscopy of membrane vesicles. Separation of the CM and the OM was attained by flotation gradient centrifugation. The appearance of the CM and OM preparations was the same in all analyzed strains—SCS1, SCS1 (RP4), and SCS1 (pML123, pWP471)—and that of the SCS1 control strain is shown. The intermediate band (marked MCM), shown for RP4 cells, contains material of both CM and OM origin. (B) The density profile (open circles) and the protein content (closed symbols) of the gradient fractions are shown. Closed circles, SCS1 closed squares, SCS1 (RP4); closed triangles, SCS1(pML123, pWP471).
FIG. 5
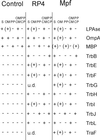
Specific release of periplasmic proteins. Control and RP4- and Mpf-containing cells were transformed to spheroplasts (S) and subsequently separated into fractions containing periplasmic proteins (PP), OM, soluble cytoplasmic (CP) and periplasmic (PP) proteins, and total membranes (OM and CM) as described in Materials and Methods. +, Mpf components immunologically identified in the periplasmic, total membrane, and soluble fractions; (+), Mpf proteins detected in small amounts in the OM. Some of the Mpf proteins were under the level of detection (u.d.) in RP4-containing cells.
FIG. 6
Immunological detection of RP4 DNA transfer and replication components in the membrane fractions. Plasmids in the SCS1 host strain encoding the indicated transfer functions or individual gene products are listed in Table 2. Flotation of the total membranes of cells containing the Tra2 region in density gradients resulted in the separation of the CM and the OM and, in addition, an intermediate band (MCM). Aliquots of the fractions, collected starting from the top of the gradient (fraction numbering is identical to that of Fig. 3), were analyzed in standard SDS-polyacrylamide gels, and proteins were transferred to PVDF membrane. Incubation with specific antisera identified the Tra1 region components (listed on the right) in different cell fractions. oe, corresponding overexpression strain; cce, crude cell extract; TM, total membranes; c, crude cell extract of the control SCS1 strain; 1 to 20, fractions collected. The arrows indicate the sample application position.
FIG. 7
RP4 DNA transfer apparatus assembly. (A) Schematic representation of individual RP4 transfer protein insertion into, association with, or translocation across the E. coli membrane. N, N terminus; C, C terminus, SEC, general secretion pathway. (B) Model of the membrane-spanning and membrane-associated DNA transfer apparatus. The size of the lettering provides a rough estimate of the relative number of molecules per Mpf complex. The spatial arrangement of TrbE, -F, -G, -I, and -L, and TraF within the complex is hypothetical. For details, see the text.
Similar articles
- TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58.
Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von Bodman SB, Lanka E, Ream W, Farrand SK. Hamilton CM, et al. J Bacteriol. 2000 Mar;182(6):1541-8. doi: 10.1128/JB.182.6.1541-1548.2000. J Bacteriol. 2000. PMID: 10692358 Free PMC article. - The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation.
Lessl M, Balzer D, Weyrauch K, Lanka E. Lessl M, et al. J Bacteriol. 1993 Oct;175(20):6415-25. doi: 10.1128/jb.175.20.6415-6425.1993. J Bacteriol. 1993. PMID: 8407818 Free PMC article. - Analysis of the sequence and gene products of the transfer region of the F sex factor.
Frost LS, Ippen-Ihler K, Skurray RA. Frost LS, et al. Microbiol Rev. 1994 Jun;58(2):162-210. doi: 10.1128/mr.58.2.162-210.1994. Microbiol Rev. 1994. PMID: 7915817 Free PMC article. Review. - The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA.
Schröder G, Lanka E. Schröder G, et al. Plasmid. 2005 Jul;54(1):1-25. doi: 10.1016/j.plasmid.2005.02.001. Plasmid. 2005. PMID: 15907535 Review.
Cited by
- Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes.
Sakai D, Komano T. Sakai D, et al. J Bacteriol. 2002 Jan;184(2):444-51. doi: 10.1128/JB.184.2.444-451.2002. J Bacteriol. 2002. PMID: 11751821 Free PMC article. - Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems.
Christie PJ. Christie PJ. Biochim Biophys Acta. 2004 Nov 11;1694(1-3):219-34. doi: 10.1016/j.bbamcr.2004.02.013. Biochim Biophys Acta. 2004. PMID: 15546668 Free PMC article. Review. - The structural biology of type IV secretion systems.
Fronzes R, Christie PJ, Waksman G. Fronzes R, et al. Nat Rev Microbiol. 2009 Oct;7(10):703-14. doi: 10.1038/nrmicro2218. Nat Rev Microbiol. 2009. PMID: 19756009 Free PMC article. Review. - Efficient markerless integration of genes in the chromosome of probiotic E. coli Nissle 1917 by bacterial conjugation.
Seco EM, Fernández LÁ. Seco EM, et al. Microb Biotechnol. 2022 May;15(5):1374-1391. doi: 10.1111/1751-7915.13967. Epub 2021 Nov 9. Microb Biotechnol. 2022. PMID: 34755474 Free PMC article. - Conjugative plasmid transfer in gram-positive bacteria.
Grohmann E, Muth G, Espinosa M. Grohmann E, et al. Microbiol Mol Biol Rev. 2003 Jun;67(2):277-301, table of contents. doi: 10.1128/MMBR.67.2.277-301.2003. Microbiol Mol Biol Rev. 2003. PMID: 12794193 Free PMC article. Review.
References
- Alt-Mörbe J, Stryker J L, Fuqua C, Li P-L, Farrand S K, Winans S C. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmid is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J Bacteriol. 1996;178:4248–4257. - PMC - PubMed
- Balzer D. Lokalisierung, Nukleotidsequenz und Genorganisation von IncP-Transferregionen. Berlin, Germany: Freie Universität Berlin; 1993.
- Bamford D H, Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi 6. Virology. 1980;107:222–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous