Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli - PubMed (original) (raw)
Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli
D E Nelson et al. J Bacteriol. 2000 Mar.
Abstract
Although general physiological functions have been ascribed to the high-molecular-weight penicillin binding proteins (PBPs) of Escherichia coli, the low-molecular-weight PBPs have no well-defined biological roles. When we examined the morphology of a set of E. coli mutants lacking multiple PBPs, we observed that strains expressing active PBP 5 produced cells of normal shape, while mutants lacking PBP 5 produced cells with altered diameters, contours, and topological features. These morphological effects were visible in untreated cells, but the defects were exacerbated in cells forced to filament by inactivation of PBP 3 or FtsZ. After filamentation, cellular diameter varied erratically along the length of individual filaments and many filaments exhibited extensive branching. Also, in general, the mean diameter of cells lacking PBP 5 was significantly increased compared to that of cells from isogenic strains expressing active PBP 5. Expression of cloned PBP 5 reversed the effects observed in DeltadacA mutants. Although deletion of PBP 5 was required for these phenotypes, the absence of additional PBPs magnified the effects. The greatest morphological alterations required that at least three PBPs in addition to PBP 5 be deleted from a single strain. In the extreme cases in which six or seven PBPs were deleted from a single mutant, cells and cell filaments expressing PBP 5 retained a normal morphology but cells and filaments lacking PBP 5 were aberrant. In no case did mutation of another PBP produce the same drastic morphological effects. We conclude that among the low-molecular-weight PBPs, PBP 5 plays a principle role in determining cell diameter, surface uniformity, and overall topology of the peptidoglycan sacculus.
Figures
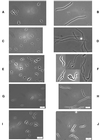
FIG. 1
Morphology of PBP mutants before and after filamentation by aztreonam. Bacteria were grown in LB broth at 37°C until reaching an _A_550 of approximately 0.2, at which point 10 μg of aztreonam per ml was added. Samples were prepared for microscopy immediately before and 45 min after addition of the antibiotic. (A) CS109; (B) CS109 plus aztreonam; (C) CS604-2; (D) CS604-2 plus aztreonam; (E) CS701-1; (F) CS701-1 plus aztreonam; (G) CS601-3; (H) CS601-3 plus aztreonam; (I) CS612-1; (J) CS612-1 plus aztreonam.
FIG. 2
Morphology of untreated PBP mutants. Bacteria were grown in LB broth at 37°C until reaching log phase and then photographed. (A) CS502-1; (B) CS508-1; (C) CS403-3; (D) CS506-1; (E) CS509-3.
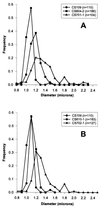
FIG. 3
Differences in cellular diameter of strains isogenic for dacA. Strains were grown in LB broth at 37°C until reaching an _A_550 of approximately 0.2, at which point samples were prepared for microscopy. “Frequency” indicates the proportion of the population with a particular diameter. n, the number of cells measured for each population.
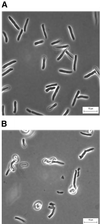
FIG. 4
Restoration of normal morphology by cloned PBP 5. E. coli CS701-1 pPJ5C was grown at 37°C in LB broth supplemented with 20 μg of chloramphenicol and either 0.005% arabinose or 0.1% glucose until reaching an _A_550 of 0.2, at which point 10 μg of aztreonam per ml was added. Samples were prepared for microscopy 45 min after addition of the antibiotic. (A) PBP 5 expression induced by 0.005% arabinose; (B) inhibition of PBP 5 expression by 0.1% glucose.
Similar articles
- Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli.
Nelson DE, Young KD. Nelson DE, et al. J Bacteriol. 2001 May;183(10):3055-64. doi: 10.1128/JB.183.10.3055-3064.2001. J Bacteriol. 2001. PMID: 11325933 Free PMC article. - Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis.
Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Denome SA, et al. J Bacteriol. 1999 Jul;181(13):3981-93. doi: 10.1128/JB.181.13.3981-3993.1999. J Bacteriol. 1999. PMID: 10383966 Free PMC article. - Sequences near the active site in chimeric penicillin binding proteins 5 and 6 affect uniform morphology of Escherichia coli.
Ghosh AS, Young KD. Ghosh AS, et al. J Bacteriol. 2003 Apr;185(7):2178-86. doi: 10.1128/JB.185.7.2178-2186.2003. J Bacteriol. 2003. PMID: 12644487 Free PMC article. - Approaching the physiological functions of penicillin-binding proteins in Escherichia coli.
Young KD. Young KD. Biochimie. 2001 Jan;83(1):99-102. doi: 10.1016/s0300-9084(00)01205-0. Biochimie. 2001. PMID: 11254981 Review. - Multiple mechanisms of membrane anchoring of Escherichia coli penicillin-binding proteins.
Gittins JR, Phoenix DA, Pratt JM. Gittins JR, et al. FEMS Microbiol Rev. 1994 Jan;13(1):1-12. doi: 10.1111/j.1574-6976.1994.tb00031.x. FEMS Microbiol Rev. 1994. PMID: 8117464 Review.
Cited by
- Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching.
Potluri LP, de Pedro MA, Young KD. Potluri LP, et al. Mol Microbiol. 2012 Apr;84(2):203-24. doi: 10.1111/j.1365-2958.2012.08023.x. Epub 2012 Mar 15. Mol Microbiol. 2012. PMID: 22390731 Free PMC article. - Electron beam fabrication of a microfluidic device for studying submicron-scale bacteria.
Moolman MC, Huang Z, Krishnan ST, Kerssemakers JW, Dekker NH. Moolman MC, et al. J Nanobiotechnology. 2013 Apr 10;11:12. doi: 10.1186/1477-3155-11-12. J Nanobiotechnology. 2013. PMID: 23575419 Free PMC article. - Genetic requirements for uropathogenic E. coli proliferation in the bladder cell infection cycle.
Mediati DG, Blair TA, Costas A, Monahan LG, Söderström B, Charles IG, Duggin IG. Mediati DG, et al. mSystems. 2024 Oct 22;9(10):e0038724. doi: 10.1128/msystems.00387-24. Epub 2024 Sep 17. mSystems. 2024. PMID: 39287381 Free PMC article. - Controlling the shape of filamentous cells of Escherichia coli.
Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Takeuchi S, et al. Nano Lett. 2005 Sep;5(9):1819-23. doi: 10.1021/nl0507360. Nano Lett. 2005. PMID: 16159230 Free PMC article. - Boosting Secretion of Extracellular Protein by Escherichia coli via Cell Wall Perturbation.
Yang H, Lu X, Hu J, Chen Y, Shen W, Liu L. Yang H, et al. Appl Environ Microbiol. 2018 Oct 1;84(20):e01382-18. doi: 10.1128/AEM.01382-18. Print 2018 Oct 15. Appl Environ Microbiol. 2018. PMID: 30097440 Free PMC article.
References
- Åkerlund T, Nordström K, Bernander R. Branched Escherichia coli cells. Mol Microbiol. 1993;10:849–858. - PubMed
- Edwards D H, Donachie W D. Construction of a triple deletion of penicillin-binding proteins 4, 5, and 6 in Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 369–374.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous