Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli - PubMed (original) (raw)
Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli
J R Johnson et al. Clin Diagn Lab Immunol. 2000 Mar.
Abstract
Repetitive-element PCR (rep-PCR) fingerprinting is a promising molecular typing tool for Escherichia coli, including for discriminating between pathogenic and nonpathogenic clones, but is plagued by irreproducibility. Using the ERIC2 and BOXA1R primers and 15 E. coli strains from the ECOR reference collection (three from each phylogenetic group, as defined by multilocus enzyme electrophoresis [MLEE], including virulence-associated group B2), we rigorously assessed the effect of extremely elevated annealing temperatures on rep-PCR's reproducibility, discriminating power, and ability to reveal MLEE-defined phylogenetic relationships. Modified cycling conditions significantly improved assay reproducibility and discriminating power, allowing fingerprints from different cyclers to be analyzed together with minimal loss of resolution. The correspondence of rep-PCR with MLEE with respect to tree structure and regression analysis of distances was substantially better with modified than with standard cycling conditions. Nonetheless, rep-PCR was only a fair surrogate for MLEE, and when fingerprints from different days were compared, it failed to distinguish between different clones within all-important phylogenetic group B2. These findings indicate that although the performance and phylogenetic fidelity of rep-PCR fingerprinting can be improved substantially with modified assay conditions, even when so improved rep-PCR cannot fully substitute for MLEE as a phylogenetic typing method for pathogenic E. coli.
Figures
FIG. 1
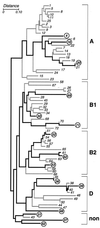
MLEE-based dendrogram for the ECOR reference collection of E. coli as derived by Herzer et al., using the NJ method to compare electrophoretic polymorphisms for 38 metabolic enzymes (23). Brackets at the right demarcate the five phylogenetic groups (A, B1, B, D, and nonaligned [non]). Heavy lines connect the 15 strains used in the present study (circles).
FIG. 2
Representative rep-PCR gels for the 15 ECOR strains, showing fingerprints as generated using the BOXA1R primer (left) or the ERIC2 primer (right), with standard cycling (top) or 65-TD cycling (bottom). Lanes are labeled with strain numbers and bracketed according to ECOR group (A, B1, B2, D, and nonaligned [non]). Lanes M, 250-bp molecular size marker. Note that fingerprints for a given strain as generated with the same primer differ considerably between standard cycling and 65-TD cycling.
FIG. 3
Single-run dendrograms for the 15 ECOR strains based on ERIC2 fingerprints (left) or BOXA1R-ERIC2 composite fingerprints (right), with 65-TD cycling. In both dendrograms, each strain is separated from all other strains. (BOXA1R fingerprints yielded similar results [data not shown].) The ERIC2 dendrogram includes both the largest (B2 strains versus others) and the smallest (B2 strains versus one another) between-strain distances observed in any of the dendrograms. non, nonaligned strains.
FIG. 4
Triplicate-run dendrograms for the 15 ECOR strains based on ERIC2 fingerprints (upper panels) or BOXA1R-ERIC2 composite fingerprints (lower panels), as generated using standard cycling (left panels) or 65-TD cycling (right panels) on cycler B. The black squares at the right of each dendrogram mark the position in the dendrogram of replicate fingerprints of each strain, as identified at the top. Solid boxes enclose fully resolved strains. Dashed boxes enclose fully resolved phylogenetic groups. (Gel strips are reconstructions and hence underestimate the clarity of the actual gel images.) Summary data for the numbers of strains and phylogenetic groups resolved are shown in Tables 4 and 5, respectively. non, nonaligned strains.
Similar articles
- Improved repetitive-element PCR fingerprinting of Salmonella enterica with the use of extremely elevated annealing temperatures.
Johnson JR, Clabots C. Johnson JR, et al. Clin Diagn Lab Immunol. 2000 Mar;7(2):258-64. doi: 10.1128/CDLI.7.2.258-264.2000. Clin Diagn Lab Immunol. 2000. PMID: 10702502 Free PMC article. - Semi-automated rep-PCR for rapid differentiation of major clonal groups of Escherichia coli meningitis strains.
Bonacorsi S, Bidet P, Mahjoub F, Mariani-Kurkdjian P, Ait-Ifrane S, Courroux C, Bingen E. Bonacorsi S, et al. Int J Med Microbiol. 2009 Aug;299(6):402-9. doi: 10.1016/j.ijmm.2009.04.001. Epub 2009 May 17. Int J Med Microbiol. 2009. PMID: 19451030 - Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting.
Johnson JR, Clabots C, Azar M, Boxrud DJ, Besser JM, Thurn JR. Johnson JR, et al. J Clin Microbiol. 2001 Oct;39(10):3452-60. doi: 10.1128/JCM.39.10.3452-3460.2001. J Clin Microbiol. 2001. PMID: 11574555 Free PMC article. - Optimization of analytical parameters for inferring relationships among Escherichia coli isolates from repetitive-element PCR by maximizing correspondence with multilocus sequence typing data.
Goldberg TL, Gillespie TR, Singer RS. Goldberg TL, et al. Appl Environ Microbiol. 2006 Sep;72(9):6049-52. doi: 10.1128/AEM.00355-06. Appl Environ Microbiol. 2006. PMID: 16957228 Free PMC article.
Cited by
- A Novel High-Resolving Method for Genomic PCR-Fingerprinting of Enterobacteria.
Isaeva AS, Kulikov EE, Tarasyan KK, Letarov AV. Isaeva AS, et al. Acta Naturae. 2010 Apr;2(1):82-8. Acta Naturae. 2010. PMID: 22649631 Free PMC article. - Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR.
Meacham KJ, Zhang L, Foxman B, Bauer RJ, Marrs CF. Meacham KJ, et al. J Clin Microbiol. 2003 Nov;41(11):5224-6. doi: 10.1128/JCM.41.11.5224-5226.2003. J Clin Microbiol. 2003. PMID: 14605168 Free PMC article. - Emergence of Citrobacter freundii carrying IMP-8 metallo-β-lactamase in Germany.
Peter S, Wolz C, Kaase M, Marschal M, Schulte B, Vogel W, Autenrieth I, Willmann M. Peter S, et al. New Microbes New Infect. 2014 Mar;2(2):42-5. doi: 10.1002/nmi2.36. Epub 2014 Mar 20. New Microbes New Infect. 2014. PMID: 25356340 Free PMC article. - The changing prevalence of drug-resistant Escherichia coli clonal groups in a community: evidence for community outbreaks of urinary tract infections.
Manges AR, Natarajan P, Solberg OD, Dietrich PS, Riley LW. Manges AR, et al. Epidemiol Infect. 2006 Apr;134(2):425-31. doi: 10.1017/S0950268805005005. Epidemiol Infect. 2006. PMID: 16490149 Free PMC article. - Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains.
Clermont O, Cordevant C, Bonacorsi S, Marecat A, Lange M, Bingen E. Clermont O, et al. J Clin Microbiol. 2001 Dec;39(12):4549-53. doi: 10.1128/JCM.39.12.4549-4553.2001. J Clin Microbiol. 2001. PMID: 11724881 Free PMC article.
References
- Arbeit R D, Maslow J N, Mulligan M E. Polymerase chain reaction-mediated genotyping in microbial epidemiology. Clin Infect Dis. 1994;18:1018–1019. - PubMed
- Berg D E, Akopyants N S, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources