Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process - PubMed (original) (raw)
Review
Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process
C R Woese et al. Microbiol Mol Biol Rev. 2000 Mar.
Abstract
The aminoacyl-tRNA synthetases (AARSs) and their relationship to the genetic code are examined from the evolutionary perspective. Despite a loose correlation between codon assignments and AARS evolutionary relationships, the code is far too highly structured to have been ordered merely through the evolutionary wanderings of these enzymes. Nevertheless, the AARSs are very informative about the evolutionary process. Examination of the phylogenetic trees for each of the AARSs reveals the following. (i) Their evolutionary relationships mostly conform to established organismal phylogeny: a strong distinction exists between bacterial- and archaeal-type AARSs. (ii) Although the evolutionary profiles of the individual AARSs might be expected to be similar in general respects, they are not. It is argued that these differences in profiles reflect the stages in the evolutionary process when the taxonomic distributions of the individual AARSs became fixed, not the nature of the individual enzymes. (iii) Horizontal transfer of AARS genes between Bacteria and Archaea is asymmetric: transfer of archaeal AARSs to the Bacteria is more prevalent than the reverse, which is seen only for the "gemini group. " (iv) The most far-ranging transfers of AARS genes have tended to occur in the distant evolutionary past, before or during formation of the primary organismal domains. These findings are also used to refine the theory that at the evolutionary stage represented by the root of the universal phylogenetic tree, cells were far more primitive than their modern counterparts and thus exchanged genetic material in far less restricted ways, in effect evolving in a communal sense.
Figures
FIG. 1
Mechanisms of aminoacyl-tRNA formation. Both pathways, direct acylation and tRNA-dependent amino acid modification, are depicted for glutaminyl-tRNA formation. For example, E. coli uses glutaminyl-tRNA synthetase while B. subtilis employs Glu-tRNAGln amidotransferase for this purpose.
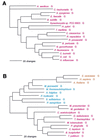
FIG. 2
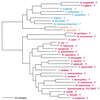
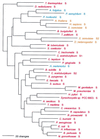
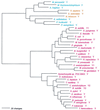
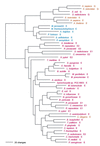
Phylogenetic tree of PheRS sequences. Aligned protein sequences were evaluated for the 1,000 most parsimonious trees (61), using amino acid replacement costs based on the BLOSUM 45 matrix (31, 41). Of these trees, we retained the topology with the maximum likelihood of giving rise to the data under the JTT model in protml version 2.2 of the MOLPHY package (1). Additional optimization was performed by removing sequences, one or more at a time, from the tree and using maximum likelihood to select the best from among the 100 to 500 most parsimonious alternative placements and to assign lengths to the branches. The tree has been rooted between the Bacteria and the Archaea plus Eukarya. The sequence identifiers correspond to an organism defined in Table 2, followed by the one-letter amino acid code (in this case F). Sometimes suffixes 1 and 2 are used; they refer to the fact that the given organism contains more than one specific AARS. Bacteria are shown in red, Archaea in blue, and eukaryotes in yellow.
FIG. 3
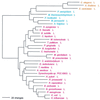
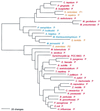
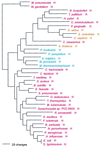
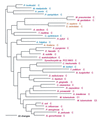
Phylogenetic tree of LeuRS sequences. The tree was rooted using ValRS, IleRS, and MetRS sequences. Other details are as in Fig. 2.
FIG. 4
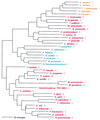
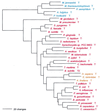
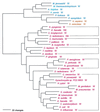
Phylogenetic tree of IleRS sequences. The tree has been rooted using ValRS, LeuRS, and MetRS sequences. Other details are as in Fig. 2.
FIG. 5
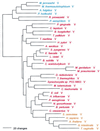
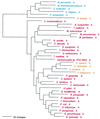
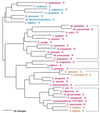
Phylogenetic tree of MetRS sequences. The tree has been rooted using LeuRS, IleRS, and ValRS sequences. Other details are as in Fig. 2.
FIG. 6
Phylogenetic tree of ValRS sequences. The tree was rooted using IleRS, MetRS, and LeuRS sequences. Other details are as in Fig. 2.
FIG. 7
Phylogenetic tree of SerRS sequences. The tree has been rooted using ThrRS sequences. The unusual SerRSs of the methanogens, which may not be related to other SerRSs, have not been used in constructing the figure. Other details are as in Fig. 2.
FIG. 8
Phylogenetic tree of ProRS sequences. The tree was rooted using ThrRS, HisRS, and GlyRS sequences. Other details are as in Fig. 2.
FIG. 9
Phylogenetic tree of ThrRS sequences. The tree has been rooted using ProRS, GlyRS, and HisRS sequences. Other details are as in Fig. 2.
FIG. 10
Phylogenetic tree of AlaRS sequences. The tree has been rooted between the bacterial sequences and the archaeal (plus Giardia) sequences. Other details are as in Fig. 2.
FIG. 11
Phylogenetic tree of TyrRS sequences. The tree has been rooted using TrpRS sequences. Other details are as in Fig. 2.
FIG. 12
Phylogenetic tree of HisRS sequences. The tree has been rooted using ThrRS, ProRS, and GlyRS sequences. Other details are as in Fig. 2.
FIG. 13
Phylogenetic tree of GlnRS sequences. The tree has been rooted using GluRS sequences. Other details are as in Fig. 2.
FIG. 14
Phylogenetic tree of AsnRS sequences. The tree has been rooted using AspRS sequences. Saccharomyces cerevisiae N1 is the cytoplasmic enzyme, while S. cerevisiae N2 is the mitochondrial enzyme. Other details are as in Fig. 2.
FIG. 15
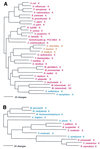
Phylogenetic trees of LysRS sequences. (A) The class II synthetases were rooted using AspRS sequences. (B) The class I (archaeal type) sequences were rooted between the crenarchaeal (plus Pyrococcus) and other euryarchaeal sequences. Other details are as in Fig. 2.
FIG. 16
Phylogenetic tree of AspRS sequences. The origin of the AsnRS sequences is also shown (designated by the N rather than the D suffixes). The tree was rooted using LysRS sequences. Other details are as in Fig. 2.
FIG. 17
Phylogenetic tree of GluRS sequences. The tree was rooted between the Bacteria and the Archaea plus Eukarya. The M. thermoautotrophicum ΔH sequence was used. Other details are as in Fig. 2.
FIG. 18
Phylogenetic tree of CysRS sequences. The tree was rooted between the largest group of archaeal sequences and all others. Other details are as in Fig. 2.
FIG. 19
Phylogenetic tree of TrpRS sequences. The tree was rooted using TyrRS sequences. Other details are as in Fig. 2. The Deinococcus radiodurans TrpRS denoted by an asterisk is chimeric (see the text). Brackets denote bacterial subtypes defined by sequence signature analysis.
FIG. 20
Phylogenetic tree of ArgRS sequences. The tree was rooted between the archaeal group and the other sequences. The S. cerevisiae M sequence is the mitochondrial enzyme. Other details are as in Fig. 2. Brackets denote bacterial subtypes defined by sequence signature analysis.
FIG. 21
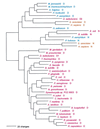
Phylogenetic tree of GlyRS sequences. (A) The α2β2 type GlyRS sequences (combining data from both subunits) were rooted on the sequence from Aquifex. (B) The homodimeric GlyRS sequences were rooted using ProRS, HisRS, and ThrRS sequences. Other details are as in Fig. 2.
Similar articles
- Enzymic recognition of amino acids drove the evolution of primordial genetic codes.
Douglas J, Bouckaert R, Carter CW Jr, Wills PR. Douglas J, et al. Nucleic Acids Res. 2024 Jan 25;52(2):558-571. doi: 10.1093/nar/gkad1160. Nucleic Acids Res. 2024. PMID: 38048305 Free PMC article. - Complex Genomes of Early Nucleocytoviruses Revealed by Ancient Origins of Viral Aminoacyl-tRNA Synthetases.
Kijima S, Hikida H, Delmont TO, Gaïa M, Ogata H. Kijima S, et al. Mol Biol Evol. 2024 Aug 2;41(8):msae149. doi: 10.1093/molbev/msae149. Mol Biol Evol. 2024. PMID: 39099254 Free PMC article. - Aminoacyl-tRNA Synthetases in the Bacterial World.
Giegé R, Springer M. Giegé R, et al. EcoSal Plus. 2016 May;7(1):10.1128/ecosalplus.ESP-0002-2016. doi: 10.1128/ecosalplus.ESP-0002-2016. EcoSal Plus. 2016. PMID: 27223819 Free PMC article. Review. - The Roots of Genetic Coding in Aminoacyl-tRNA Synthetase Duality.
Carter CW Jr, Wills PR. Carter CW Jr, et al. Annu Rev Biochem. 2021 Jun 20;90:349-373. doi: 10.1146/annurev-biochem-071620-021218. Epub 2021 Mar 29. Annu Rev Biochem. 2021. PMID: 33781075 Review.
Cited by
- Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Kuhle B, Hirschi M, Doerfel LK, Lander GC, Schimmel P. Kuhle B, et al. Nat Commun. 2022 Aug 30;13(1):5100. doi: 10.1038/s41467-022-32544-1. Nat Commun. 2022. PMID: 36042193 Free PMC article. - Emergence and evolution.
Bullwinkle TJ, Ibba M. Bullwinkle TJ, et al. Top Curr Chem. 2014;344:43-87. doi: 10.1007/128_2013_423. Top Curr Chem. 2014. PMID: 23478877 Free PMC article. Review. - Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1.
Akochy PM, Bernard D, Roy PH, Lapointe J. Akochy PM, et al. J Bacteriol. 2004 Feb;186(3):767-76. doi: 10.1128/JB.186.3.767-776.2004. J Bacteriol. 2004. PMID: 14729703 Free PMC article. - Editorial: Noncanonical functions of Aminoacyl-tRNA synthetases.
Sun L, Zhou XL, Zhou ZW, Cui H. Sun L, et al. Front Physiol. 2023 Feb 23;14:1165515. doi: 10.3389/fphys.2023.1165515. eCollection 2023. Front Physiol. 2023. PMID: 36909230 Free PMC article. No abstract available. - On the evolution of cells.
Woese CR. Woese CR. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8742-7. doi: 10.1073/pnas.132266999. Epub 2002 Jun 19. Proc Natl Acad Sci U S A. 2002. PMID: 12077305 Free PMC article.
References
- Adduce J, Hoosegow M. MOLPHY: programs for maximum likelihood inference of protein phylogeny. Computer science monographs, no. 27. Tokyo, Japan: Institute of Statistical Mathematics; 1992.
- Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
- Arnez J G, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–216. - PubMed
- Berthet-Colominas C, Seignovert L, Härtlein M, Grotli M, Cusack S, Leberman R. The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J. 1998;17:2947–2960. - PMC - PubMed
- Brown J R. Aminoacyl-tRNA synthetases: evolution of a troubled family. In: Wiegel J, Adams M H W, editors. Thermophiles: the keys to molecular evolution and the origin of life? London, United Kingdom: Taylor & Francis; 1998. pp. 217–230.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous