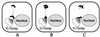Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly - PubMed (original) (raw)
Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly
A G Bost et al. J Virol. 2000 Apr.
Abstract
The replicase gene (gene 1) of the coronavirus mouse hepatitis virus (MHV) encodes two co-amino-terminal polyproteins presumed to incorporate all the virus-encoded proteins necessary for viral RNA synthesis. The polyproteins are cotranslationally processed by viral proteinases into at least 15 mature proteins, including four predicted cleavage products of less than 25 kDa that together would comprise the final 59 kDa of protein translated from open reading frame 1a. Monospecific antibodies directed against the four distinct domains detected proteins of 10, 12, and 15 kDa (p1a-10, p1a-12, and p1a-15) in MHV-A59-infected DBT cells, in addition to a previously identified 22-kDa protein (p1a-22). When infected cells were probed by immunofluorescence laser confocal microscopy, p1a-10, -22, -12, and -15 were detected in discrete foci that were prominent in the perinuclear region but were widely distributed throughout the cytoplasm as well. Dual-labeling experiments demonstrated colocalization of the majority of p1a-22 in replication complexes with the helicase, nucleocapsid, and 3C-like proteinase, as well as with p1a-10, -12, and -15. p1a-22 was also detected in separate foci adjacent to the replication complexes. The majority of complexes containing the gene 1 proteins were distinct from sites of accumulation of the M assembly protein. However, in perinuclear regions the gene 1 proteins and nucleocapsid were intercalated with sites of M protein localization. These results demonstrate that the complexes known to be involved in RNA synthesis contain multiple gene 1 proteins and are closely associated with structural proteins at presumed sites of virion assembly.
Figures
FIG. 1
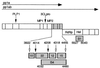
MHV-A59 gene 1 organization, putative cleavage sites, and cloned protein domains. The organization and arrangement of protein domains comprising polyprotein 1a (pp1a, ORF1a) and polyprotein 1ab (pp1ab, ORF1a/b) are shown. The locations of the papain-like proteinase 1 (PLP1) and 3CLpro are indicated by arrows above the schematic, and the predicted or confirmed cleavage products are demarcated by vertical bars. The location of the membrane protein domains (MP1 and MP2) in pp1a and the RNA-dependent RNA polymerase (RdRp) and helicase (Hel) in pp1ab are also shown. The cassette of four proteins in the carboxy-terminal portion of pp1a is indicated in gray. In the lower schematic, the p1a-10, -22, -12, and -15 protein domains are shown enlarged with filled arrows denoting MHV-A59 3CLpro cleavage sites that have been confirmed in vitro and open arrows indicating predicted 3CLpro cleavage sites. The cloned fragments used for production of antisera against the mature proteins (αp1a-10, -22, -12, and -15, designated by mass in kilodaltons) or against a fusion protein spanning the p1a-22 through p1a-15 domains (the B4 antibody) are also shown in gray. The fusion protein used for generating the B1 (α-helicase) antibody is similarly indicated. Relevant MHV-A59 amino acid numbers at the termini of each protein product are noted.
FIG. 2
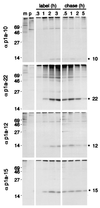
Identification of proteins processed from the carboxy-terminal portion of the ORF1a polyprotein in MHV-A59-infected cells. MHV-A59-infected (i) or mock-infected (m) DBT cell lysates were immunoprecipitated with the antisera directed against the individual protein domains as indicated above each lane and analyzed by SDS–10 to 20% gradient polyacrylamide gel electrophoresis and fluorography. Molecular mass markers (kilodaltons) are shown to the left of the gel.
FIG. 3
Pulse-label and pulse-chase translation of p1a-10, -22, -12, and -15. For pulse-label translation (label), DBT cells were mock infected (m) or infected with MHV-A59 for 5.5 h and were incubated with [35S]Cys-Met for 0.3, 1, 2, or 3 h prior to immunoprecipitation with the antibodies as indicated to the right of each gel. p denotes immunoprecipitation with preimmune serum. For pulse-chase translation (chase), DBT cells were radiolabeled beginning at 5.5 h p.i. with [35S]Cys-Met for 90 min prior to addition of excess cold methionine and cycloheximide for the times indicated above each well. Following harvesting and immunoprecipitation of the cells, label and chase samples for each antibody were run on the same SDS–10 to 20% gradient polyacrylamide gel, followed by fluorography. Molecular mass markers (kilodaltons) are shown to the left of the gels, and the locations of the p1a-10, -22, -12, and -15 proteins are indicated to the right of the gels.
FIG. 4
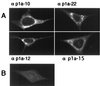
Localization of p1a-10, -22, -12, and -15 in MHV-infected cells. MHV-A59-infected or mock-infected DBT cells were fixed and prepared for immunofluorescence microscopy as described in Materials and Methods, using the antibodies as noted by each frame. Cells were imaged on a Zeiss LSM 410 confocal microscope using a 488-nm laser with acquisition in the LSM software. The images are single confocal slices using a 63× objective. Image processing (brightness and contrast) was performed in Photoshop 5.0. A representative mock-infected cell probed with αp1a-22 is shown in panel B. αp1a-10, -12, and -15 resulted in a similar lack of background staining in the mock-infected cells.
FIG. 5
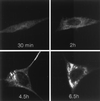
Time course of p1a-22 localization during MHV-A59 infection. Infected DBT cells were fixed in methanol at the times p.i. as indicated prior to preparation for immunofluorescence using αp1a-22. Confocal images were acquired and modified as described for Fig. 4. All images were identically processed to allow direct comparison of the extent of p1a-22 expression.
FIG. 6
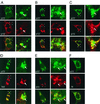
Dual-labeling immunofluorescence confocal microscopy of MHV-infected cells. At 6.5 h p.i. MHV-infected DBT cells were fixed and prepared for immunofluorescence using αp1a-22 as a primary antibody and other gene 1 protein antibodies or the αN monoclonal antibody as the second primary antibody. p1a-22 (p22) is shown in green in each panel, and the other proteins are shown in red as indicated. The merged images are shown with areas of colocalization in yellow. The right column of each panel is a higher magnification (×5.3) of the area demarcated by the white boxes in the left image. Arrows indicate areas of intercalation of p1a-22 with the other gene 1 proteins. (A) p1a-10 (p10); (B) p1a-12 (p12); (C) p1a-15 (p15); (D) helicase (hel); (E) 3CLpro (pro); (F) nucleocapsid (N).
FIG. 7
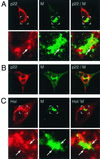
Dual-labeling imaging of M and p1a-22 or helicase. DBT cells were infected for 6 h prior to fixation and preparation for dual-labeling immunofluorescence using αM (green in all images) and either αp1a-22 (p22) (A and B) or B1 αhel (Hel) (C). (A and C) The bottom panels show higher magnifications of the region marked by the white box in the corresponding upper panel. Arrows indicate regions of intercalation of p1a-22 or hel with M. (B) Localization of p1a-22 and M in a virus-induced syncytium.
FIG. 8
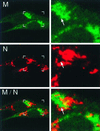
M and N are distinct but closely associated in perinuclear regions. At 6 h p.i., infected BHK-R cells were fixed and labeled with αN (J.3.3) followed by Cy3-conjugated anti-mouse secondary antibody and Cy2-conjugated αM (J.1.3). In the left panel are shown two infected cells with boxes corresponding to the increased-magnification panels to the right. Arrows indicate areas of intercalation between M and nucleocapsid (N). Areas of colocalization are shown in yellow.
FIG. 9
Possible models for interdigitation of replication complexes with sites of virion assembly. (A) Replication complexes throughout the cytoplasm (black) move along cytoskeletal pathways to sites juxtaposed to IC-Golgi membranes. (B) RNA-nucleocapsids move from replication complexes (dark grey) via separate transport complexes (black). (C) Only amplified or newly formed replication complexes (black) directly adjacent to the IC-Golgi membranes interact with sites of assembly. The model is not intended to describe the source of membranes for replication complexes.
Similar articles
- The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis.
Denison MR, Spaan WJ, van der Meer Y, Gibson CA, Sims AC, Prentice E, Lu XT. Denison MR, et al. J Virol. 1999 Aug;73(8):6862-71. doi: 10.1128/JVI.73.8.6862-6871.1999. J Virol. 1999. PMID: 10400784 Free PMC article. - Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection.
Bost AG, Prentice E, Denison MR. Bost AG, et al. Virology. 2001 Jun 20;285(1):21-9. doi: 10.1006/viro.2001.0932. Virology. 2001. PMID: 11414802 Free PMC article. - Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes.
Sims AC, Ostermann J, Denison MR. Sims AC, et al. J Virol. 2000 Jun;74(12):5647-54. doi: 10.1128/jvi.74.12.5647-5654.2000. J Virol. 2000. PMID: 10823872 Free PMC article. - Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro.
Lu XT, Sims AC, Denison MR. Lu XT, et al. J Virol. 1998 Mar;72(3):2265-71. doi: 10.1128/JVI.72.3.2265-2271.1998. J Virol. 1998. PMID: 9499085 Free PMC article. - The nucleocapsid of vesicular stomatitis virus.
Luo M. Luo M. Sci China Life Sci. 2012 Apr;55(4):291-300. doi: 10.1007/s11427-012-4307-x. Epub 2012 May 9. Sci China Life Sci. 2012. PMID: 22566085 Review.
Cited by
- Studying the dynamics of coronavirus replicative structures.
Hagemeijer MC, de Haan CA. Hagemeijer MC, et al. Methods Mol Biol. 2015;1282:261-9. doi: 10.1007/978-1-4939-2438-7_22. Methods Mol Biol. 2015. PMID: 25720487 Free PMC article. - The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing.
Snijder EJ, Decroly E, Ziebuhr J. Snijder EJ, et al. Adv Virus Res. 2016;96:59-126. doi: 10.1016/bs.aivir.2016.08.008. Epub 2016 Sep 14. Adv Virus Res. 2016. PMID: 27712628 Free PMC article. Review. - Expression, crystallization and preliminary crystallographic study of mouse hepatitis virus (MHV) nucleocapsid protein C-terminal domain.
Tong X, Ma Y, Li X. Tong X, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Jun 1;66(Pt 6):674-6. doi: 10.1107/S1744309110012492. Epub 2010 May 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20516597 Free PMC article. - Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region.
Spencer KA, Hiscox JA. Spencer KA, et al. FEBS Lett. 2006 Oct 30;580(25):5993-8. doi: 10.1016/j.febslet.2006.09.052. Epub 2006 Oct 2. FEBS Lett. 2006. PMID: 17052713 Free PMC article. - Expression, purification, and characterization of SARS coronavirus RNA polymerase.
Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, Sarafianos SG, Ding J. Cheng A, et al. Virology. 2005 May 10;335(2):165-76. doi: 10.1016/j.virol.2005.02.017. Virology. 2005. PMID: 15840516 Free PMC article.
References
- Bi W, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Intracellular localization of polypeptides encoded in mouse hepatitis virus open reading frame 1A. Adv Exp Med Biol. 1995;380:251–258. - PubMed
- Bi W, Pinon J D, Hughes S, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J Neurovirol. 1998;4:594–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI026603/AI/NIAID NIH HHS/United States
- AI-26603/AI/NIAID NIH HHS/United States
- AI01479/AI/NIAID NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- IP30CA68485/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources