Glucose depletion rapidly inhibits translation initiation in yeast - PubMed (original) (raw)
Glucose depletion rapidly inhibits translation initiation in yeast
M P Ashe et al. Mol Biol Cell. 2000 Mar.
Free PMC article
Abstract
Glucose performs key functions as a signaling molecule in the yeast Saccharomyces cerevisiae. Glucose depletion is known to regulate gene expression via pathways that lead to derepression of genes at the transcriptional level. In this study, we have investigated the effect of glucose depletion on protein synthesis. We discovered that glucose withdrawal from the growth medium led to a rapid inhibition of protein synthesis and that this effect was readily reversed upon readdition of glucose. Neither the inhibition nor the reactivation of translation required new transcription. This inhibition also did not require activation of the amino acid starvation pathway or inactivation of the TOR kinase pathway. However, mutants in the glucose repression (reg1, glc7, hxk2, and ssn6), hexose transporter induction (snf3 rgt2), and cAMP-dependent protein kinase (tpk1(w) and tpk2(w)) pathways were resistant to the inhibitory effects of glucose withdrawal on translation. These findings highlight the intimate connection between the nutrient status of the cell and its translational capacity. They also help to define a new area of posttranscriptional regulation in yeast.
Figures
Figure 1
Summary of the signaling pathways involving translational or glucose-mediated controls in S. cerevisiae. Arrows represent activating signals, and blunt-ended lines represent inhibitory signals.
Figure 2
Glucose or fructose removal from yeast medium specifically inhibits translation initiation. (A) Polyribosome traces from the wild-type strain (yAS2568). Yeast was grown in YPD and resuspended in YP medium lacking (YP) or containing (YPD) glucose for the indicated times (minutes). Polyribosomes were analyzed as described in MATERIALS AND METHODS. The peaks that contain the small ribosomal subunit (40S), the large ribosomal subunit (60S), and both subunits (80S) are indicated by arrows. The polysome peaks generated by 2, 3, 4, 5, etc. 80S ribosomes on a single mRNA are bracketed. (B) Polyribosome traces from wild-type yeast that were washed in the absence of glucose (YP) for 10 min and then incubated in medium containing glucose for the indicated times (minutes). (C) Polyribosome traces from wild-type yeast grown on YP medium with a variety of 2% carbon sources (fructose [F], sucrose [S], maltose [M], and raffinose [R]). Cells were washed for 10 min either in the presence (+) or absence (−) of the carbon source. (D) [35S]Methionine incorporation into proteins over time in wild-type yeast. Cells were grown in synthetic complete minus methionine medium, harvested, and resuspended in a labeling mixture in the presence (♦) or absence (□) of a carbon source (glucose [D] or galactose [G]). Aliquots were taken at the indicated times, and the levels of [35S]methionine incorporated into proteins were determined. (E) Polyribosome traces from wild-type yeast grown in galactose medium (YPG) and resuspended in YP or YPG medium for the indicated times (minutes).
Figure 3
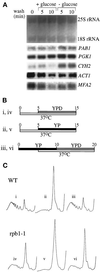
Neither mRNA degradation nor transcription contributes to the inhibition of translation upon glucose withdrawal or the recovery after glucose readdition. (A) Northern blot stained with methylene blue to visualize the rRNA levels (upper panel). The same blot was probed for a variety of specific mRNAs (lower panels) (see MATERIALS AND METHODS for details). (B) Line diagram for the experimental protocol used in C. The Roman numerals refer to the polyribosome traces shown in C. The timing (minutes) and length of the temperature change from 30 to 37°C is depicted by the lower, white bars. The upper bars represent medium changes and length of washes (gray bars represent a change to YPD, and black bars represent a change to YP). (C) Polyribosome traces for the wild-type (yAS306) and RNA polymerase II mutant rpb1-1 (yAS879) strains. Roman numerals refer to B, which depicts the order and timing of YP or YPD washes and temperature changes that occurred before cells were harvested for polyribosome analysis.
Figure 4
Mutants in other translational control pathways are still inhibited for translation upon glucose withdrawal. (A) Polyribosome traces from the wild-type (yAS2570) and SUI2-S51A (yAS2571) strains. Yeast was grown in SCD-Leu medium and washed for 10 min in SCD-Leu (+D), SC-Leu (−D), or SCD minus all amino acids (−AAs). (B) Polyribosome traces from the wild-type (yAS2410) and tap42-11 (yAS2411) strains. Yeast was grown in SCD-Leu medium and then washed for 10 min in either SCD-Leu (+D) or SC-Leu (−D).
Figure 5
Polyribosome analyses for a selection of strains used in this study. Examples of polyribosome traces generated with the use of the wild-type strain (yAS2572 [FY250]) and various mutant strains (yAS2576 [_reg1Δ_], yAS2605 [_reg1Δ snf1Δ_], yAS2578 [_hxk2Δ_], yAS2606 [_hxk2Δ snf1Δ_], yAS2498 [_ssn6Δ_], yAS2537 [_rgt2Δ snf3Δ_], yAS2602 [_rgt2Δ snf3Δ snf1Δ_], yAS963 [tpk1 w1 _tpk2Δ tpk3Δ_], yAS964 [tpk1Δ tpk2 w1 _tpk3Δ_], yAS2599 [_tpk1Δ tpk2Δ tpk3Δ msn2Δ msn4Δ_], and yAS2577 [_grr1Δ_]) washed for 10 min in the presence (+D) or absence (−D) of glucose.
Figure 6
Measurement of intracellular ATP levels after glucose withdrawal from the growth medium. The percentage of the starting intracellular ATP level for 30 min after transfer of yeast to medium with glucose (□) or without glucose (♦) is plotted. The W3031A gal graph is identical to the others except that the strain was grown in galactose medium and was transferred to medium with (□) or without (♦) galactose. Strains used are the wild-type strains yAS2572 (FY250), yAS962 (SP1), and yAS2568 (W3031A) and the mutant strains yAS2578 (hxk2Δ), yAS2576 (reg1Δ), yAS2577 (grr1Δ), and yAS963 (tpk1 w1 tpk2Δ tpk3Δ).
Figure 7
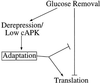
Multiple pathways may be involved in glucose-dependent translational control. Glucose removal is shown to inhibit translation. Translation resumes after switching carbon sources via a process termed adaptation. Adaptation might either directly reverse the translational inhibition or somehow overcome the inhibition by activating translation. Adaptation appears to be induced by the derepression of glucose-repressible genes and/or by low cAPK activity (both of which are also brought about by glucose removal).
Similar articles
- Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae.
Slattery MG, Liko D, Heideman W. Slattery MG, et al. Eukaryot Cell. 2008 Feb;7(2):358-67. doi: 10.1128/EC.00334-07. Epub 2007 Dec 21. Eukaryot Cell. 2008. PMID: 18156291 Free PMC article. - Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2.
Conlan RS, Tzamarias D. Conlan RS, et al. J Mol Biol. 2001 Jun 22;309(5):1007-15. doi: 10.1006/jmbi.2001.4742. J Mol Biol. 2001. PMID: 11399075 - Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose.
Ozcan S, Johnston M. Ozcan S, et al. Mol Cell Biol. 1995 Mar;15(3):1564-72. doi: 10.1128/MCB.15.3.1564. Mol Cell Biol. 1995. PMID: 7862149 Free PMC article. - How do yeast cells sense glucose?
Kruckeberg AL, Walsh MC, Van Dam K. Kruckeberg AL, et al. Bioessays. 1998 Dec;20(12):972-6. doi: 10.1002/(SICI)1521-1878(199812)20:12<972::AID-BIES2>3.0.CO;2-M. Bioessays. 1998. PMID: 10048296 Review. - Glucose-sensing and -signalling mechanisms in yeast.
Rolland F, Winderickx J, Thevelein JM. Rolland F, et al. FEMS Yeast Res. 2002 May;2(2):183-201. doi: 10.1111/j.1567-1364.2002.tb00084.x. FEMS Yeast Res. 2002. PMID: 12702307 Review.
Cited by
- Characterization of Arabidopsis thaliana GCN2 kinase roles in seed germination and plant development.
Liu X, Merchant A, Rockett KS, McCormack M, Pajerowska-Mukhtar KM. Liu X, et al. Plant Signal Behav. 2015;10(4):e992264. doi: 10.4161/15592324.2014.992264. Plant Signal Behav. 2015. PMID: 25912940 Free PMC article. - Timing and Variability of Galactose Metabolic Gene Activation Depend on the Rate of Environmental Change.
Nguyen-Huu TD, Gupta C, Ma B, Ott W, Josić K, Bennett MR. Nguyen-Huu TD, et al. PLoS Comput Biol. 2015 Jul 22;11(7):e1004399. doi: 10.1371/journal.pcbi.1004399. eCollection 2015 Jul. PLoS Comput Biol. 2015. PMID: 26200924 Free PMC article. - The ribosome-associated chaperone Zuo1 controls translation upon TORC1 inhibition.
Black A, Williams TD, Soubigou F, Joshua IM, Zhou H, Lamoliatte F, Rousseau A. Black A, et al. EMBO J. 2023 Dec 11;42(24):e113240. doi: 10.15252/embj.2022113240. Epub 2023 Nov 20. EMBO J. 2023. PMID: 37984430 Free PMC article. - Novel activity of eukaryotic translocase, eEF2: dissociation of the 80S ribosome into subunits with ATP but not with GTP.
Demeshkina N, Hirokawa G, Kaji A, Kaji H. Demeshkina N, et al. Nucleic Acids Res. 2007;35(14):4597-607. doi: 10.1093/nar/gkm468. Epub 2007 Jun 22. Nucleic Acids Res. 2007. PMID: 17586816 Free PMC article. - Identification of a 57S translation complex containing closed-loop factors and the 60S ribosome subunit.
Denis CL, Laue TM, Wang X. Denis CL, et al. Sci Rep. 2018 Jul 31;8(1):11468. doi: 10.1038/s41598-018-29832-6. Sci Rep. 2018. PMID: 30065356 Free PMC article.
References
- Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases