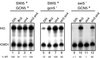Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription - PubMed (original) (raw)
Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription
Y Yu et al. Mol Cell Biol. 2000 Apr.
Abstract
Recent work has shown that transcription of the yeast HO gene involves the sequential recruitment of a series of transcription factors. We have performed a functional analysis of HO regulation by determining the ability of mutations in SIN1, SIN3, RPD3, and SIN4 negative regulators to permit HO expression in the absence of certain activators. Mutations in the SIN1 (=SPT2) gene do not affect HO regulation, in contrast to results of other studies using an HO:lacZ reporter, and our data show that the regulatory properties of an HO:lacZ reporter differ from that of the native HO gene. Mutations in SIN3 and RPD3, which encode components of a histone deacetylase complex, show the same pattern of genetic suppression, and this suppression pattern differs from that seen in a sin4 mutant. The Sin4 protein is present in two transcriptional regulatory complexes, the RNA polymerase II holoenzyme/mediator and the SAGA histone acetylase complex. Our genetic analysis allows us to conclude that Swi/Snf chromatin remodeling complex has multiple roles in HO activation, and the data suggest that the ability of the SBF transcription factor to bind to the HO promoter may be affected by the acetylation state of the HO promoter. We also demonstrate that the Nhp6 architectural transcription factor, encoded by the redundant NHP6A and NHP6B genes, is required for HO expression. Suppression analysis with sin3, rpd3, and sin4 mutations suggests that Nhp6 and Gcn5 have similar functions. A gcn5 nhp6a nhp6b triple mutant is extremely sick, suggesting that the SAGA complex and the Nhp6 architectural transcription factors function in parallel pathways to activate transcription. We find that disruption of SIN4 allows this strain to grow at a reasonable rate, indicating a critical role for Sin4 in detecting structural changes in chromatin mediated by Gcn5 and Nhp6. These studies underscore the critical role of chromatin structure in regulating HO gene expression.
Figures
FIG. 1
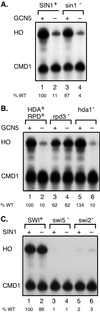
HO expression is not altered by a sin1 mutation. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1 in each panel. RNAs were prepared from strains DY2395, DY5116, DY5323, and DY5326 (A), DY2389, DY5199, DY4548, DY5170, DY5068, and DY5168 (B), and DY150, DY5323, DY161, DY5410, DY2348, and DY5420 (C).
FIG. 2
The nhp6 defect in HO transcription can be suppressed by sin3 or sin4 mutations. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1. RNAs were prepared from strains DY150, DY2381, DY3658, DY5153, DY984, DY5157, DY1699, and DY5155.
FIG. 3
Both sin3 and sin4 mutations suppress the gcn5 defect. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1. RNAs were prepared from strains DY150, DY5285, DY2763, DY5294, DY5265, DY5297, DY5289, DY5315, DY411, DY775, DY2133, and DY5299.
FIG. 4
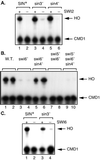
A sin4 mutation suppresses swi6 but not swi2. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). (A) HO is not expressed in swi2 sin3 or swi2 sin4 strains. RNAs were prepared from strains DY150, DY3944, DY773, DY2870, DY1702, and DY2499. (B) HO is expressed in swi6 sin4 strains. RNAs were prepared from strains DY150, DY151, DY5780, DY5781, DY5907, DY5908, DY5909, DY5910, DY5911, and DY5912. (C) HO is not expressed in a swi6 sin3 strains. RNAs were prepared from strains DY150, DY5780, DY773, and DY6103.
FIG. 5
Nhp6b overexpression partially suppresses the gcn5 defect. Strains DY150 (wild type [w.t.]), DY161 (swi5), DY2348 (swi2), DY5116 (gcn5), and DY5780 (swi6) were transformed with either the YEplac195 vector or M1195, a YEp-NHP6B plasmid. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control), using RNA prepared from strains grown under selective conditions to maintain the plasmid. The upper panel was exposed to film for 8 h; the lower panel was exposed for 24 h.
FIG. 6
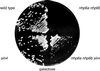
A sin4 mutation suppresses the nhp6 growth defect on galactose. Strains DY881, DY1712, DY2532, and DY2533 were plated on YEP-galactose medium and grown for 4 days at 30°C.
FIG. 7
The severe growth defect of a gcn5 nhp6a nhp6b triple mutant is suppressed by a sin4 mutation. (A) Strains DY150, DY2378, DY2380, DY2382, DY5116, and DY5306 were plated on YEPD medium and grown for 3 days at 30°C. (B) Strains DY5825 and DY5820 were plated on YEPD medium and grown for 5 days at 30°C.
Similar articles
- SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment.
Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. Mitra D, et al. Mol Cell Biol. 2006 Jun;26(11):4095-110. doi: 10.1128/MCB.01849-05. Mol Cell Biol. 2006. PMID: 16705163 Free PMC article. - Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.
Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Biswas D, et al. Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article. - Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein.
Yu Y, Eriksson P, Bhoite LT, Stillman DJ. Yu Y, et al. Mol Cell Biol. 2003 Mar;23(6):1910-21. doi: 10.1128/MCB.23.6.1910-1921.2003. Mol Cell Biol. 2003. PMID: 12612066 Free PMC article. - A SAGA of histone acetylation and gene expression.
Hampsey M. Hampsey M. Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
- Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae.
Stillman DJ. Stillman DJ. Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):175-80. doi: 10.1016/j.bbagrm.2009.11.010. Biochim Biophys Acta. 2010. PMID: 20123079 Free PMC article. Review. - The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes.
Biddick RK, Law GL, Chin KK, Young ET. Biddick RK, et al. J Biol Chem. 2008 Nov 28;283(48):33101-9. doi: 10.1074/jbc.M805258200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826948 Free PMC article. - SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment.
Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. Mitra D, et al. Mol Cell Biol. 2006 Jun;26(11):4095-110. doi: 10.1128/MCB.01849-05. Mol Cell Biol. 2006. PMID: 16705163 Free PMC article. - Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.
Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Biswas D, et al. Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article. - Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae.
Biswas D, Yu Y, Mitra D, Stillman DJ. Biswas D, et al. Genetics. 2006 Feb;172(2):837-49. doi: 10.1534/genetics.105.050245. Epub 2005 Nov 4. Genetics. 2006. PMID: 16272410 Free PMC article.
References
- Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. - PubMed
- Björklund S, Almouzni G, Davidson I, Nightingale K P, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases