Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation - PubMed (original) (raw)
Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation
H Hirochika et al. Plant Cell. 2000 Mar.
Abstract
Gene silencing associated with repeated DNA sequences has been reported for many eukaryotes, including plants. However, its biological significance remains to be determined. One important function that has been proposed is the suppression of transposons. Here, we address transposon suppression by examining the behavior of the tobacco retrotransposon Tto1 and endogenous retrotransposons in Arabidopsis. After an initial increase in copy number because of active transposition in the Arabidopsis genome, Tto1 became silent. The amount of transcript was reduced, and the inactivated Tto1 became methylated. This silencing correlated with an increase in copy number. These phenomena mimic repeat-induced gene silencing. The homozygous ddm1 (for decrease in DNA methylation) mutation of Arabidopsis results in genomic DNA hypomethylation and the release of silencing in repeated genes. To investigate the role of DNA methylation and the gene-silencing machinery in the suppression of Tto1, we introduced the ddm1 mutation into an Arabidopsis line carrying inactivated Tto1 copies. In the homozygous ddm1 background, Tto1 became hypomethylated and transcriptionally and transpositionally active. In addition, one of the newly isolated endogenous Arabidopsis retrotransposon families, named Tar17, also became hypomethylated and transcriptionally active in the ddm1 mutant background. Our results suggest that the inactivation of retrotransposons and the silencing of repeated genes have mechanisms in common.
Figures
Figure 1.
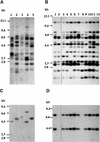
DNA Gel Blot Analysis of Transposition of Tto1 in Transgenic Arabidopsis Lines. (A) and (C) Analyses of five independent T0 transgenic lines. (B) and (D) Analyses of the T1 progeny of one T0 line (lane 4 in [A]). Genomic DNA was digested with EcoRV and hybridized with a _Tto1_-gag probe ([A] and [B]) or an HPT probe ([C] and [D]). DNA length markers are shown at left in kilobases.
Figure 2.
PCR Analysis of Transposition of Tto1 in T0 Transgenic Lines. (A) Process of transposition and recovery of the 5′ deleted sequence expected from transposition. A derivative of Tto1-1 carrying a 36-bp deletion at the 5′ end (_Tto1_[−36]) was inserted into the T-DNA region of the vector plasmid, resulting in pBITto1(−36). In transgenic lines, the 5′ deleted sequence is expected to be recovered during transposition by means of an RNA. A 630-bp fragment was amplified only from transposed Tto1 copies by using primers 1 and 2, because primer 1 is homologous to the deleted sequence. LTR, long terminal repeat. (B) Results of PCR analysis. Five T0 transgenic lines used in Figure 1A and control transgenic lines transformed with the vector plasmid (pBI101-Hm) were subjected to PCR analysis. The plasmid carrying Tto1(−36), pSKTto1(−36), was also used as a control. M, HincII-digested φX174 as a length marker.
Figure 3.
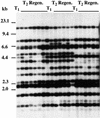
Effect of Tissue Culture on Transposition in the Progeny of Transgenic Lines. Hypocotyl sections of pooled progeny (10 T2 plants from each T1 plant) of three T1 plants derived from one T0 line (Figure 1B, lanes 2, 4, and 5) were cultured on callus-inducing medium. Genomic DNA prepared from T1 plants (T1) and from regenerated plants (T2 Regen.) from induced calli was digested with EcoRV and subjected to DNA gel blot analysis with the Tto1 gag probe. DNA length markers are shown at left in kilobases.
Figure 4.
Analysis of DNA Methylation of Tto1 in T0, T1, and T2 Transgenic Lines. (A) Restriction maps of Tto1-1 and probes used for the analysis. Black rectangles denote the long terminal repeat. (B) DNA gel blot analysis of methylation of Tto1 in transgenic lines having high copy numbers. Genomic DNA was digested with HpaII or MspI and analyzed by DNA gel blotting with the Tto1 gag probe. T2 DNA was prepared from the pooled progeny (four regenerated T2 plants from each T1 plant used in Figure 3). T1 DNA was prepared from the pooled progeny (five T1 plants) derived from each T0 line. (C) DNA gel blot analysis of methylation of Tto1 in transgenic lines having low or medium copy numbers. Genomic DNA was digested with HpaII and analyzed by DNA gel blotting with the Tto1 gag probe. T1 DNA was prepared from the pooled progeny (10 T1 plants) derived from each T0 line. The copy numbers of Tto1 in transgenic lines (from left to right) are six, two, one, and one (data not shown). Only the left-most transgenic line carries transposed copies (four copies). All of the DNA samples analyzed in (B) and (C) were shown to be equally well digested with the restriction enzyme by reprobing the blot with a single-copy sequence (m105; Pruitt and Meyerowitz, 1986) (data not shown). Lengths and positions of fragments expected from digestion of unmethylated Tto1 are shown at right in (B) and (C) in kilobases.
Figure 5.
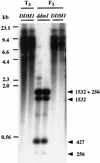
Analysis of DNA Methylation of Tto1 in the Wild Type and ddm1 Mutant. Genomic DNAs prepared from two T3 plants (T3; lines 121 and 122), and from F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families (from a cross T2 plant [_Tto1 DDM1_] × [_ddm1_]) were digested with EcoRV and analyzed by DNA gel blotting with the Tto1 gag probe. All of the DNA samples were found to be equally well digested by the restriction enzyme by reprobing the blot with the single-copy sequence discussed in the legend to Figure 4 (data not shown). DNA length markers are shown at left in kilobases. Expected lengths of fragments are shown at right in kilobases.
Figure 6.
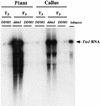
Analysis of Tto1 RNA in the Wild Type and ddm1 Mutant. Total RNA was prepared from whole plants or calli of T3 plants (T3; lines 121 and 122) and F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families. As a control, total RNA from tobacco BY2 cells (Nagata et al., 1981) was analyzed. Twenty micrograms of total RNA was loaded on the gel.
Figure 7.
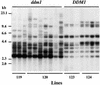
Reactivation of Tto1 Transposition by the ddm1 Mutation in Calli. Calli were induced from F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families and cultured for 3 months. Induced calli were smashed into pieces, and each piece was cultured separately for one more month. DNA from each callus was digested with EcoRV and analyzed by DNA gel blotting with the Tto1 gag probe. DNA length markers are shown at left in kilobases.
Figure 8.
Analysis of the Linear Molecule of Tto1 in the ddm1 Mutant. Total DNAs prepared from calli of F3 ddm1/ddm1 (ddm1; lines 119 and 120) and DDM1/DDM1 (DDM1; lines 123 and 124) families were analyzed without restriction digestion by DNA blotting with the Tto1-gag probe. The linear Tto1 molecule is indicated by an arrow. M, HindIII-digested λ DNA as a length marker.
Figure 9.
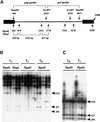
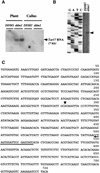
Activation of Transcription of the Endogenous Retrotransposon Tar17 by the ddm1 Mutation. (A) RNA gel blot analysis. Total RNAs prepared from whole plants or calli of ddm1/ddm1 (ddm1) and DDM1/DDM1 (DDM1) were analyzed by using the Tto1 gag probe. (B) Primer extension analysis. Ten micrograms of total RNA extracted from whole plants was hybridized with the 5′ end-labeled oligonucleotide. The hybrids were extended with reverse transcriptase, and the cDNA products were electrophoresed on a sequencing gel alongside a sequencing reaction using the same primer, as described previously (Hirochika, 1993). One band (indicated by an arrowhead) was detected in the ddm1 mutant only. (C) Nucleotide sequence of the LTR. The 5′ end of Tar17 RNA, determined by primer extension, is indicated by an arrowhead. The position of the oligonucleotide used for primer extension analysis is indicated by an arrow.
Similar articles
- Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1.
Saze H, Kakutani T. Saze H, et al. EMBO J. 2007 Aug 8;26(15):3641-52. doi: 10.1038/sj.emboj.7601788. Epub 2007 Jul 12. EMBO J. 2007. PMID: 17627280 Free PMC article. - Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis.
Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Miura A, et al. Nature. 2001 May 10;411(6834):212-4. doi: 10.1038/35075612. Nature. 2001. PMID: 11346800 - Linear DNA intermediates of the Tto1 retrotransposon in Gag particles accumulated in stressed tobacco and Arabidopsis thaliana.
Takeda S, Sugimoto K, Kakutani T, Hirochika H. Takeda S, et al. Plant J. 2001 Nov;28(3):307-17. doi: 10.1046/j.1365-313x.2001.01151.x. Plant J. 2001. PMID: 11722773 - Regulation of retrotransposition in Arabidopsis.
Lee SC, Martienssen RA. Lee SC, et al. Biochem Soc Trans. 2021 Nov 1;49(5):2241-2251. doi: 10.1042/BST20210337. Biochem Soc Trans. 2021. PMID: 34495315 Review. - Transcriptional gene silencing mutants.
Mittelsten Scheid O, Paszkowski J. Mittelsten Scheid O, et al. Plant Mol Biol. 2000 Jun;43(2-3):235-41. doi: 10.1023/a:1006487529698. Plant Mol Biol. 2000. PMID: 10999407 Review.
Cited by
- Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Osakabe A, Takizawa Y, Horikoshi N, Hatazawa S, Negishi L, Sato S, Berger F, Kakutani T, Kurumizaka H. Osakabe A, et al. Nat Commun. 2024 Jul 11;15(1):5187. doi: 10.1038/s41467-024-49465-w. Nat Commun. 2024. PMID: 38992002 Free PMC article. - Epigenetic weapons of plants against fungal pathogens.
Mierziak J, Wojtasik W. Mierziak J, et al. BMC Plant Biol. 2024 Mar 6;24(1):175. doi: 10.1186/s12870-024-04829-8. BMC Plant Biol. 2024. PMID: 38443788 Free PMC article. Review. - Transposable elements: multifunctional players in the plant genome.
Hassan AH, Mokhtar MM, El Allali A. Hassan AH, et al. Front Plant Sci. 2024 Jan 4;14:1330127. doi: 10.3389/fpls.2023.1330127. eCollection 2023. Front Plant Sci. 2024. PMID: 38239225 Free PMC article. Review. - Two ARGONAUTE proteins loaded with transposon-derived small RNAs are associated with the reproductive cell lineage in Arabidopsis.
Bradamante G, Nguyen VH, Incarbone M, Meir Z, Bente H, Donà M, Lettner N, Scheid OM, Gutzat R. Bradamante G, et al. Plant Cell. 2024 Mar 29;36(4):863-880. doi: 10.1093/plcell/koad295. Plant Cell. 2024. PMID: 38060984 Free PMC article. - From parasites to partners: exploring the intricacies of host-transposon dynamics and coevolution.
Chakrabarty P, Sen R, Sengupta S. Chakrabarty P, et al. Funct Integr Genomics. 2023 Aug 23;23(3):278. doi: 10.1007/s10142-023-01206-w. Funct Integr Genomics. 2023. PMID: 37610667 Review.
References
- Akama, K., Shiraishi, H., Ohta, S., Nakamura, K., Okada, K., and Shimura, Y. (1992). Efficient transformation of Arabidopsis thaliana: Comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 12 7–11. - PubMed
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279 2113–2115. - PubMed
- Assaad, F.F., Tucker, K.L., and Signer, E.R. (1993). Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22 1067–1085. - PubMed
- Bender, J., and Fink, G.R. (1995). Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83 725–734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources