Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures - PubMed (original) (raw)
Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures
S Krause et al. Proc Natl Acad Sci U S A. 2000.
Abstract
RP4 TrbB, an essential component of the conjugative transfer apparatus of the broad-host-range plasmid RP4, is a member of the PulE protein superfamily involved in multicomponent machineries transporting macromolecules across the bacterial envelope. PulE-like proteins share several well conserved motifs, most notable a nucleoside triphosphate binding motif (P-loop). Helicobacter pylori HP0525 also belongs to the PulE superfamily and is encoded by the pathogenicity island cag, involved in the inflammatory response of infected gastric epithelial cells in mammals. The native molecular masses of TrbB and HP0525 as determined by gel filtration and glycerol gradient centrifugation suggested a homohexameric structure in the presence of ATP and Mg(2+). In the absence of nucleotides and bivalent cations, TrbB behaved as a tetramer whereas the hexameric state of HP0525 remained unaffected. Electron microscopy and image processing demonstrated that TrbB and HP0525 form ring-shaped complexes (diameter: 12 nm) with a central region (diameter: 3 nm) of low electron density when incubated in the presence of ATP and Mg(2+). However, the TrbB average image appeared to be more elliptical with strong twofold rotational symmetry whereas HP0525 complexes are regular hexagons. Six well defined triangle-shaped areas of high electron density were distinguishable in both cases. Covalent crosslinking of TrbB suggests that the hexameric ring is composed from a trimer of dimers, because only dimeric, tetrameric, and hexameric species were detectable. The toroidal structure of TrbB and HP0525 suggests that both proteins catalyze a repetitive process, most probably translocating a cognate substrate across the inner membrane.
Figures
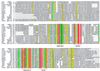
Figure 1
Alignment of VirB11 family transport NTPases. Amino acid sequences were aligned by using the
pileup
program of the GCG program package version 9.1.
boxshade
3.31 (
http://www.ch.embnet.org/software/BOX\_form.html
) was used with the following parameters: red background and white letters, 100% conservation; green background and white letters, at least 75% conservation of identical residue; and yellow background, at least 75% conservation of similar residues. The extensions of the four common motifs are marked by horizontal lines. Accession numbers: TraG (pKM101), I79275; TrwD(R388), X81123; VirB11 (pTi15955), X06826; VirB11 (pTiC58), P07169; VirB11 (Rhizobium etli), AF176227; VirB11 (Bartonella henselae), AF182718; PtlH (B. pertussis), F47301; VirB11 (Brucella suis), AF141604; RP292 (Rickettsia prowazekii), AJ235271; HP0525 (Helicobacter pylori cag), AE00148; TrbB (RP4), M93696; TrbB (R751), U67194; TrbB (pTiA6NC), P54907; TrbB (Rhizobium sp. NGR234), AE000068; Kbh (pCl1 Chlorobium limicula), U77780; and VirB11 (pXO1 Bacillus anthracis), AF065404.
Figure 2
Glycerol gradient centrifugation of TrbB and HP0525. Purified TrbB (A) [fraction IV, 150 μl, 435 μg; (20)] and purified HP0525 (B) [fraction IV, 150 μl, 435 μg; (20)] were laid on a 3.7-ml, 15–35% (wt/vol) linear glycerol gradient in buffer E or F. Centrifugation was at 270,000 × g for 15 h at 4°C. Before centrifugation, TrbB and HP0525 were incubated in buffer F for 10 min at 30°C. Fractions were tested for dATPase activity as published elsewhere (20, 30). Aliquots were electrophoresed in SDS/15% polyacrylamide gels. Gels were stained with Serva Blue R and scanned in the Personal Densitometer (Molecular Dynamics). The amount of protein was quantified by using the
imagequant
software. The stained gels of the glycerol gradient centrifugation in the absence of dATP and Mg2+ are shown in a (TrbB) and d (HP0525). Corresponding graphs are shown above the gels. Gels c (TrbB) and e (HP0525) are from glycerol gradient centrifugation in the presence of dATP and Mg2+. Corresponding graphs are shown below the gels. TrbB was also incubated with dADP and Mg2+, and glycerol gradient centrifugation was carried out in the presence of dADP and Mg2+ (b). In all graphs shown, open squares represent the ATPase activity of the fractions, filled squares, the amount of protein. Scales are given on the left and right hand sides of the graphs. Catalase (1: 240 kDa, _s_20,w = 11.3 S), aldolase (2: 185 kDa, _s_20,w = 7.8 S), and BSA (3: 68 kDa, _s_20,w = 4.4 S) were run in parallel as standards and marked by arrows.
Figure 3
Covalent crosslinking of TrbB and HP0525 subunits. TrbB protein (5 μM) and 15 μM HP0525, respectively, were incubated with various concentrations of glutaraldehyde in buffer G at 25°C for 90 min. Reactions were stopped with 3.5 M urea (final concentration). Denatured samples were electrophoresed in an SDS/10% polyacrylamide gel. Gels were stained with Serva Blue R. (A) Crosslinking of TrbB. Lanes: a, molecular mass markers; b, no glutaraldehyde; c, 100 μM; and d, 300 μM glutaraldehyde, respectively. (B) Crosslinking of HP0525. Lanes: a, molecular mass markers; b, no glutaraldehyde; c, 650 μM; and d, 1,300 μM glutaraldehyde, respectively.
Figure 4
Typical electron micrographs of negatively stained TrbB (a) and HP0525 (b) samples in the presence of Mg2+ and ATP. The arrrowheads point to some characteristic particles selected from these fields. The bar represents 50 nm. Two galleries with the type of images that were used in the two-dimensional analysis are shown in c (TrbB) and d (HP0525). The global average images are shown in e (TrbB) and f (HP0525) and have been low-pass filtered to their final resolution (14 Å in both cases). The size of the frames of each image in the galleries and for the final average images is 20.0 × 20.0 nm.
Similar articles
- The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption.
Rabel C, Grahn AM, Lurz R, Lanka E. Rabel C, et al. J Bacteriol. 2003 Feb;185(3):1045-58. doi: 10.1128/JB.185.3.1045-1058.2003. J Bacteriol. 2003. PMID: 12533481 Free PMC article. - TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates?
Schröder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. Schröder G, et al. J Bacteriol. 2002 May;184(10):2767-79. doi: 10.1128/JB.184.10.2767-2779.2002. J Bacteriol. 2002. PMID: 11976307 Free PMC article. - Helicobacter pylori CagA: analysis of sequence diversity in relation to phosphorylation motifs and implications for the role of CagA as a virulence factor.
Evans DJ Jr, Evans DG. Evans DJ Jr, et al. Helicobacter. 2001 Sep;6(3):187-98. doi: 10.1046/j.1523-5378.2001.00028.x. Helicobacter. 2001. PMID: 11683921 Review. - Analysis of the sequence and gene products of the transfer region of the F sex factor.
Frost LS, Ippen-Ihler K, Skurray RA. Frost LS, et al. Microbiol Rev. 1994 Jun;58(2):162-210. doi: 10.1128/mr.58.2.162-210.1994. Microbiol Rev. 1994. PMID: 7915817 Free PMC article. Review.
Cited by
- Protein-protein interactions among Helicobacter pylori cag proteins.
Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. Busler VJ, et al. J Bacteriol. 2006 Jul;188(13):4787-800. doi: 10.1128/JB.00066-06. J Bacteriol. 2006. PMID: 16788188 Free PMC article. - Conjugative plasmid transfer in gram-positive bacteria.
Grohmann E, Muth G, Espinosa M. Grohmann E, et al. Microbiol Mol Biol Rev. 2003 Jun;67(2):277-301, table of contents. doi: 10.1128/MMBR.67.2.277-301.2003. Microbiol Mol Biol Rev. 2003. PMID: 12794193 Free PMC article. Review. - Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism.
Yamagata A, Tainer JA. Yamagata A, et al. EMBO J. 2007 Feb 7;26(3):878-90. doi: 10.1038/sj.emboj.7601544. Epub 2007 Jan 25. EMBO J. 2007. PMID: 17255937 Free PMC article. - Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27.
Rose I, Biuković G, Aderhold P, Müller V, Grüber G, Averhoff B. Rose I, et al. Extremophiles. 2011 Mar;15(2):191-202. doi: 10.1007/s00792-010-0343-2. Epub 2011 Jan 6. Extremophiles. 2011. PMID: 21210168 - Biological diversity of prokaryotic type IV secretion systems.
Alvarez-Martinez CE, Christie PJ. Alvarez-Martinez CE, et al. Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review.
References
- Pansegrau W, Lanka E. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. - PubMed
- Lessl M, Balzer D, Pansegrau W, Lanka E. J Biol Chem. 1992;267:20471–20480. - PubMed
- Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. J Mol Biol. 1994;239:623–663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources