Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote - PubMed (original) (raw)
Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote
L Garczarek et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Two ecotypes of the prokaryote Prochlorococcus adapted to distinct light niches in the ocean have been described recently. These ecotypes are characterized by their different (divinyl-) chlorophyll (Chl) a to Chl b ratios and 16S rRNA gene signatures, as well as by their significantly distinct irradiance optima for growth and photosynthesis [Moore, L. R., Rocap, G. & Chisholm, S. W. (1998) Nature (London) 393, 464-467]. However, the molecular basis of their physiological differences remained, so far, unexplained. In this paper, we show that the low-light-adapted Prochlorococcus strain SS120 possesses a gene family of seven transcribed genes encoding different Chl a/b-binding proteins (Pcbs). In contrast, Prochlorococcus sp. MED4, a high-light-adapted ecotype, possesses a single pcb gene. The presence of multiple antenna genes in another low-light ecotype (NATL2a), but not in another high-light ecotype (TAK9803-2), is demonstrated. Thus, the multiplication of pcb genes appears as a key factor in the capacity of deep Prochlorococcus populations to survive at extremely low photon fluxes.
Figures
Figure 1
Alignment of the deduced protein sequences from the Prochlorococcus MED4 pcbA gene (M-PcbA) and from Prochlorococcus SS120 pcbA to pcbG genes (S-PcbA to S-PcbG). Identical residues are shown in white type on a black background. Black type on gray squares indicates that the percentage of conserved residues is >60% (i.e., at least 5 of 8 sequences). Predicted membrane-spanning regions (MSR) are indicated by arrows, and stars mark putative Chl-binding residues.
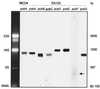
Figure 2
Detection of antenna gene transcripts by Northern blotting from Prochlorococcus MED4 (pcbA) and SS120 (pcbA to pcbG) strains. Transcripts of pcbF have been indicated by an arrow. The sizes of major RNA molecules are indicated.
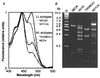
Figure 3
Characterization of different low-light (LL)- and high-light (HL)-adapted Prochlorococcus ecotypes grown at 20 μmol Q m−2⋅s−1. (A) Excitation fluorescence spectra of whole cells normalized to the divinyl-Chl a peak (444 nm). The variable height of the second peak, mainly monovinyl-Chl b (468 nm) for NATL2a and divinyl-Chl b (478 nm) for the other strains, indicates the widely different Chl b to Chl a ratios between strains. (B) Pattern of amplified pcb fragments obtained on polyacrylamide gels after cleavage by _Hae_III. Lane M, size markers.
Similar articles
- Expression and phylogeny of the multiple antenna genes of the low-light-adapted strain Prochlorococcus marinus SS120 (Oxyphotobacteria).
Garczarek L, van der Staay GW, Hess WR, Le Gall F, Partensky F. Garczarek L, et al. Plant Mol Biol. 2001 Aug;46(6):683-93. doi: 10.1023/a:1011681726654. Plant Mol Biol. 2001. PMID: 11575723 - Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem.
Bibby TS, Mary I, Nield J, Partensky F, Barber J. Bibby TS, et al. Nature. 2003 Aug 28;424(6952):1051-4. doi: 10.1038/nature01933. Nature. 2003. PMID: 12944966 - Analysis of the hli gene family in marine and freshwater cyanobacteria.
Bhaya D, Dufresne A, Vaulot D, Grossman A. Bhaya D, et al. FEMS Microbiol Lett. 2002 Oct 8;215(2):209-19. doi: 10.1111/j.1574-6968.2002.tb11393.x. FEMS Microbiol Lett. 2002. PMID: 12399037 - Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies.
Ting CS, Rocap G, King J, Chisholm SW. Ting CS, et al. Trends Microbiol. 2002 Mar;10(3):134-42. doi: 10.1016/s0966-842x(02)02319-3. Trends Microbiol. 2002. PMID: 11864823 Review. - Prochlorococcus, a marine photosynthetic prokaryote of global significance.
Partensky F, Hess WR, Vaulot D. Partensky F, et al. Microbiol Mol Biol Rev. 1999 Mar;63(1):106-27. doi: 10.1128/MMBR.63.1.106-127.1999. Microbiol Mol Biol Rev. 1999. PMID: 10066832 Free PMC article. Review.
Cited by
- Gene family size conservation is a good indicator of evolutionary rates.
Chen FC, Chen CJ, Li WH, Chuang TJ. Chen FC, et al. Mol Biol Evol. 2010 Aug;27(8):1750-8. doi: 10.1093/molbev/msq055. Epub 2010 Mar 1. Mol Biol Evol. 2010. PMID: 20194423 Free PMC article. - Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments.
García-Fernández JM, de Marsac NT, Diez J. García-Fernández JM, et al. Microbiol Mol Biol Rev. 2004 Dec;68(4):630-8. doi: 10.1128/MMBR.68.4.630-638.2004. Microbiol Mol Biol Rev. 2004. PMID: 15590777 Free PMC article. - Quantitative Proteomics Shows Extensive Remodeling Induced by Nitrogen Limitation in Prochlorococcus marinus SS120.
Domínguez-Martín MA, Gómez-Baena G, Díez J, López-Grueso MJ, Beynon RJ, García-Fernández JM. Domínguez-Martín MA, et al. mSystems. 2017 May 30;2(3):e00008-17. doi: 10.1128/mSystems.00008-17. eCollection 2017 May-Jun. mSystems. 2017. PMID: 28593196 Free PMC article. - Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp. strain PCC 9511.
Holtzendorff J, Partensky F, Jacquet S, Bruyant F, Marie D, Garczarek L, Mary I, Vaulot D, Hess WR. Holtzendorff J, et al. J Bacteriol. 2001 Feb;183(3):915-20. doi: 10.1128/JB.183.3.915-920.2001. J Bacteriol. 2001. PMID: 11208789 Free PMC article. - In vivo regulation of glutamine synthetase activity in the marine chlorophyll b-containing cyanobacterium Prochlorococcus sp. strain PCC 9511 (oxyphotobacteria).
El Alaoui S, Diez J, Humanes L, Toribio F, Partensky F, García-Fernández JM. El Alaoui S, et al. Appl Environ Microbiol. 2001 May;67(5):2202-7. doi: 10.1128/AEM.67.5.2202-2207.2001. Appl Environ Microbiol. 2001. PMID: 11319101 Free PMC article.
References
- Chisholm S W, Frankel S L, Goericke R, Olson R J, Palenik B, Waterbury J B, West-Johnsrud L, Zettler E R. Arch Microbiol. 1992;157:297–300.
- Moore L R, Rocap G, Chisholm S W. Nature (London) 1998;393:464–467. - PubMed
- Moore L R, Chisholm S W. Limnol Oceanogr. 1999;44:628–638.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases