A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge - PubMed (original) (raw)
A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge
G T Belz et al. J Virol. 2000 Apr.
Abstract
Respiratory challenge of H-2(b) mice with an H3N2 influenza A virus causes an acute, transient pneumonitis characterized by the massive infiltration of CD8(+) T lymphocytes. The inflammatory process monitored by quantitative analysis of lymphocyte populations recovered by bronchoalveolar lavage is greatly enhanced by prior exposure to an H1N1 virus, with the recall of cross-reactive CD8(+)-T-cell memory leading to more rapid clearance of the infection from the lungs. The predominant epitope recognized by the influenza virus-specific CD8(+) set has long been thought to be a nucleoprotein (NP(366-374)) presented by H-2D(b) (D(b)NP(366)). This continues to be true for the secondary H3N2-->H1N1 challenge but can no longer be considered the case for the primary response to either virus. Quantitative analysis based on intracellular staining for gamma interferon has shown that the polymerase 2 protein (PA(224-233)) provides a previously undetected epitope (D(b)PA(224)) that is at least as prominent as D(b)NP(366) during the first 10 days following primary exposure to either the H3N2 or H1N1 virus. The response to D(b)NP(366) seems to continue for longer, even when infectious virus can no longer be detected, but there is no obvious difference in the prevalence of memory T cells specific for D(b)NP(366) and D(b)PA(224). The generalization that the magnitude of the functional memory T-cell pool is a direct consequence of the clonal burst size during the primary response may no longer be useful. Previous CD8(+)-T-cell immunodominance heirarchies defined largely by cytotoxic T-lymphocyte assays may need to be revised.
Figures
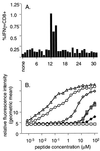
FIG. 1
Identification of the H-2Db-restricted PA224–233 (DbPA224) epitope. (A) Overlapping 15-aa PA peptides, sharing 5 aa with both the preceding and the subsequent peptides, were used to stimulate enriched CD8+ splenocytes pooled from two B6 mice at 10 days after i.n. infection with 106.8 EID50 of the HKx31 influenza A virus. The incubations were continued for 5 h in the presence of brefeldin A, and the percentage of CD8+ IFN-γ+ cells was determined by flow cytometry after surface staining for CD8, fixation, and permeabilization followed by staining for intracellular IFN-γ (Pepγ assay). (B) The epitope identified from the overlapping set responding in panel A did not conform to the H-2Db consensus motif XXXXNXXXM/I/L (5). Peptides encompassing α1 aa of the flanking regions were therefore examined for upregulation of surface expression of H-2Db on RMAS cells. The cells were incubated in titrated amounts of each peptide for 3 h and then stained for H-2Db with 28.14.85 rabbit anti-mouse–FITC and examined by flow cytometry for the geometric mean fluorescence intensity. The NP366 peptide (ASNENMETM) (▵) was used as a positive control. Data are representative of three independent experiments: ○, SSLENFRAYV; ◊, SLENFRAYV; ⧫, PSSLENFRAYV; ⊕, LENFRAYV; ⊞, SSLENFRAY.
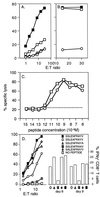
FIG. 2
Assaying functional CD8+ T cells specific for DbPA224 and variant peptides. (A) Spleen cells from B6 mice primed 2 months previously by i.n. challenge with 106.8 EID50 of the HKx31 influenza A virus were cultured for 5 days with syngeneic, PA224-pulsed stimulator cells. The lymphocytes were then assayed (6 h) on 51Cr-labeled EL4 cells (H-2b) that were pulsed with 1 μM PA224 (■) or NP366 (◊), infected for 1 h with the HKx31 virus (□), or left untreated (○). (B) Lytic activity was measured for primary CD8+ T cells stimulated with PA224-pulsed cells and then assayed on 51Cr-labeled EL4 targets that had been incubated with 10−6 M (■) or 10−9 M (□) PA224 or left untreated (○). (C) Memory CD8+ T cells from mice infected 2 months previously with HKx31 were restimulated with HKx31-infected syngeneic splenocytes for 5 days and then assayed for lytic activity against EL4 cells pulsed with log dilutions of either DbNP366 or DbPA224 peptide. ○, DbNP366; □, DbPA224 peptide; –––, unpulsed targets. (D) The lytic activity of a CD8+-T-cell line specific for DbPA224 was measured (left y axis) using EL4 target cells pulsed with 1 μM PA224 analogues in which single alanine substitutions were made at position 2, 5, 9, or 10. These peptides were used to stimulate BAL fluid cells obtained 8 or 9 days after challenge with the HKx31 virus for analysis by the Pepγ assay (right y axis).
FIG. 3
Flow cytometric assay of peptide-specific CD8+ T cells recovered directly from acutely infected mice. Freshly isolated BAL fluid (upper panel), spleen (lower left panels), and MLN (lower right panels) populations from B6 mice infected i.n. with the HKx31 influenza virus 8 days previously were cultured for 5 h in the presence or absence of viral peptide and brefeldin A, stained for CD8α, fixed and permeabilized, and then stained for intracellular IFN-γ (Pepγ assay). Adherent cells were first removed from pooled (three mice) BAL fluid suspensions by incubation on plastic for 60 min at 37°C, while CD8+ T cells were enriched from the spleen and MLN samples (see Materials and Methods). The percentages of CD8+ cells in a lymphocyte/lymphoblast gate staining for IFN-γ after incubation without peptide were 0.16, 0.14, and 0.04 for BAL fluid, spleen, and MLN populations, respectively.
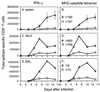
FIG. 4
Kinetic analysis of peptide-specific CD8+-T-cell responses following primary challenge with the HKx31 (H3N2) or PR8 (H1N1) influenza A virus. Naive B6 mice were infected i.n. with 106.8 EID50 of the HKx31 virus (A to C) or 20 EID50 of the PR8 virus (D to F). Spleen cells (A, D, and G) were assayed from individual mice (results are shown as mean and SE), while the populations recovered from the MLN (B, E, and H) and the BAL fluid (C and F) were pooled (n = 5 or 6). The lymphocytes were then processed, stimulated with 1 μM peptide, and stained for CD8 and IFN-γ (Pepγ assay). The percentages of CD8+ T cells specific for DbNP366 (NP), DbPA224 (PA), KbNS2114 (NS2), and KbM1128 (M1) were determined by flow cytometry (Fig. 3), and then the numbers of epitope-specific CD8+ T cells in each anatomical site were calculated using the percentage of staining and the total counts recovered. The percentages of CD8+ T cells were also analyzed statistically: the spleen results for DbNP366 and DbPA224 were significantly different (P<0.005) on day 10 and day 13 for the mice primed i.p. with PR8 (G) and on day 13 for those challenged i.n. with this virus (D).
FIG. 5
Contemporary analysis of the secondary CD8+-T-cell response using the Pepγ and tetramer-staining protocols. The B6 mice were primed by i.p. injection with 108.5 EID50 of the PR8 virus and challenged i.n. 42 days later with 106.8 EID50 of the HKx31 virus. The percentage of peptide-specific CD8+ T cells was determined by the Pepγ assay (A, C, and E) or by tetramer staining (B, D, and F). Spleen cells (A and B) were assayed from individual mice (results shown as mean and SE), while the populations recovered from the MLN (C and D) and the BAL fluid (E and F) were pooled (n = 3 to 5). The lymphocytes were then processed, stimulated with 1 μM peptide, and stained for CD8 and IFN-γ (Pepγ assay). The percentages of CD8+ T cells specific for DbNP366 (NP) (▵) and DbPA224 (PA) (○) were determined by flow cytometry, and the numbers of epitope-specific CD8+ T cells in each anatomical site were calculated using the percentage of cells staining and the total counts recovered. Estimates of memory-T-cell frequency prior to secondary challenge are also shown as ratios in panels B, D, and F, although the percentage of cells staining with the tetrameric reagents is too low for the MLN results to be meaningful. Also, although the percentage of tetramer-positive cells in the BAL fluid population is high on day 0, the numbers recovered from the lung are minuscule.
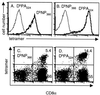
FIG. 6
Specificity of the tetrameric complexes. (A and B) Peptide-specific CD8+-T-cell lines were generated by successive rounds of bulk culture from HKx31 immune splenocytes exposed to peptide-pulsed stimulators in the presence of human rIL-2. The staining profiles for the DbNP366 (A) and DbPA224 (B) specific sets are shown for the CD3+ CD8+ lymphoblast gate. The results for unstained cells were comparable to those for the irrelevant tetrameric complex. (C and D) BAL fluid cells were obtained from B6 mice 8 days after primary infection with the HKx31 influenza virus, adhered for 1 h to remove adherent mononuclear cells, and then stained for DbNP366 (C) and DbPA224 (D).
FIG. 7
ELISpot analysis of CD8+-T-cell memory established by primary infection. The mice were infected i.p. with the PR8 virus or i.n. with the HKx31 virus and tested 42 days (PR8) or from 50 to 250 days (HKx31) later. The results show the reciprocal frequency of spot-forming cells specific for DbNP366 or DbPA224. Unfractionated cells from four or five individual spleens were incubated on IFN-γ-coated ELISpot plates with peptide-pulsed (1 μM) syngeneic spleen cells. The extent of IFN-γ secretion was determined after 44 to 48 h with a second biotinylated anti-IFN-γ MAb and streptavidin-alkaline phosphatase. The limit of detection in this assay (dotted line) was approximately 25,000 cells/peptide-specific ELISpot.
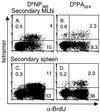
FIG. 8
Determination of cell cycling in DbNP366- and DbPA224-specific CD8+-T-cell populations during secondary influenza virus infection. Mice were primed i.p. with the PR8 virus and rested for 84 days before being infected i.n. with the HKx31 influenza virus. They were fed continuously with BrdU-containing water and analyzed 8 days after the HKx31 challenge. The MLN (A and B) and spleen (C and D) populations were enriched for CD8+ T cells and surface stained for CD8α and the relevant Db-tetramer, followed by fixation and intracellular staining for BrdU. The prevalence of the tetramer+ BrdU+ set was determined for the CD8+ population. The profiles shown are for pooled MLN from five mice and for an individual spleen.
Similar articles
- Measuring the diaspora for virus-specific CD8+ T cells.
Marshall DR, Turner SJ, Belz GT, Wingo S, Andreansky S, Sangster MY, Riberdy JM, Liu T, Tan M, Doherty PC. Marshall DR, et al. Proc Natl Acad Sci U S A. 2001 May 22;98(11):6313-8. doi: 10.1073/pnas.101132698. Epub 2001 May 8. Proc Natl Acad Sci U S A. 2001. PMID: 11344265 Free PMC article. - Antigen-specific CD8(+) T cells persist in the upper respiratory tract following influenza virus infection.
Wiley JA, Hogan RJ, Woodland DL, Harmsen AG. Wiley JA, et al. J Immunol. 2001 Sep 15;167(6):3293-9. doi: 10.4049/jimmunol.167.6.3293. J Immunol. 2001. PMID: 11544317 - Characterization of a new H-2D(k)-restricted epitope prominent in primary influenza A virus infection.
Tourdot S, Herath S, Gould KG. Tourdot S, et al. J Gen Virol. 2001 Jul;82(Pt 7):1749-1755. doi: 10.1099/0022-1317-82-7-1749. J Gen Virol. 2001. PMID: 11413387 - Effect of MHC class I diversification on influenza epitope-specific CD8+ T cell precursor frequency and subsequent effector function.
Day EB, Charlton KL, La Gruta NL, Doherty PC, Turner SJ. Day EB, et al. J Immunol. 2011 Jun 1;186(11):6319-28. doi: 10.4049/jimmunol.1000883. Epub 2011 May 2. J Immunol. 2011. PMID: 21536802 Free PMC article. - Characterization of CD8+ T cell repertoire diversity and persistence in the influenza A virus model of localized, transient infection.
Turner SJ, Kedzierska K, La Gruta NL, Webby R, Doherty PC. Turner SJ, et al. Semin Immunol. 2004 Jun;16(3):179-84. doi: 10.1016/j.smim.2004.02.005. Semin Immunol. 2004. PMID: 15130502 Review.
Cited by
- Use it or lose it: establishment and persistence of T cell memory.
Kedzierska K, Valkenburg SA, Doherty PC, Davenport MP, Venturi V. Kedzierska K, et al. Front Immunol. 2012 Nov 27;3:357. doi: 10.3389/fimmu.2012.00357. eCollection 2012. Front Immunol. 2012. PMID: 23230439 Free PMC article. - The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype.
Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. Boon AC, et al. J Virol. 2002 Jan;76(2):582-90. doi: 10.1128/jvi.76.2.582-590.2002. J Virol. 2002. PMID: 11752149 Free PMC article. - Preinduced reovirus-specific T-cell immunity enhances the anticancer efficacy of reovirus therapy.
Groeneveldt C, Kinderman P, van Stigt Thans JJC, Labrie C, Griffioen L, Sluijter M, van den Wollenberg DJM, Hoeben RC, den Haan JMM, van der Burg SH, van Hall T, van Montfoort N. Groeneveldt C, et al. J Immunother Cancer. 2022 Jul;10(7):e004464. doi: 10.1136/jitc-2021-004464. J Immunother Cancer. 2022. PMID: 35853671 Free PMC article. - A modular approach to assembly of totally synthetic self-adjuvanting lipopeptide-based vaccines allows conformational epitope building.
Zeng W, Horrocks KJ, Robevska G, Wong CY, Azzopardi K, Tauschek M, Robins-Browne RM, Jackson DC. Zeng W, et al. J Biol Chem. 2011 Apr 15;286(15):12944-51. doi: 10.1074/jbc.M111.227744. Epub 2011 Feb 14. J Biol Chem. 2011. PMID: 21321114 Free PMC article. - Chronic spinal cord injury attenuates influenza virus-specific antiviral immunity.
Bracchi-Ricard V, Zha J, Smith A, Lopez-Rodriguez DM, Bethea JR, Andreansky S. Bracchi-Ricard V, et al. J Neuroinflammation. 2016 May 31;13(1):125. doi: 10.1186/s12974-016-0574-y. J Neuroinflammation. 2016. PMID: 27245318 Free PMC article.
References
- Altman J D, Moss P, Goulder P, Barouch D H, McHeyzer-Williams M, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. - PubMed
- Busch D H, Pilip I M, Vijh S, Pamer E G. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. - PubMed
- Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous