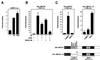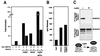Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases - PubMed (original) (raw)
Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases
J Lu et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Myocyte enhancer factor-2 (MEF2) transcription factors control muscle-specific and growth factor-inducible genes. We show that hypertrophic growth of cardiomyocytes in response to phenylephrine and serum is accompanied by activation of MEF2 through a posttranslational mechanism mediated by calcium, calmodulin-dependent protein kinase (CaMK), and mitogen-activated protein kinase (MAPK) signaling. CaMK stimulates MEF2 activity by dissociating class II histone deacetylases (HDACs) from the DNA-binding domain. MAPKs, which activate MEF2 by phosphorylation of the transcription activation domain, maximally stimulate MEF2 activity only when repression by HDACs is relieved by CaMK signaling to the DNA-binding domain. These findings identify MEF2 as an endpoint for hypertrophic stimuli in cardiomyocytes and demonstrate that MEF2 mediates synergistic transcriptional responses to the CaMK and MAPK signaling pathways by signal-dependent dissociation from HDACs.
Figures
Figure 1
CaMK and MAPK target different domains of MEF2. Primary neonatal rat cardiomyocytes in serum-free medium were transiently transfected with the indicated expression plasmids, and luciferase activity was determined in cell extracts. (A) Cells were stimulated with PE (10 μM) or 10% FBS, as indicated, and expression of the MEF2-dependent reporter, 3xMEF2-luciferase, was assayed. (B) Cells transiently transfected with pG5E1b-luciferase (Gal-luc) and GAL-MEF2C were stimulated with PE, as in A, in the presence of KN62 or SB202190, as indicated. (C) Cells were transiently transfected with pG5E1b-luciferase and GAL-MEF2C (Left) or GAL-MEF2C-ΔN (Right), along with activated CaMKIV and MKK6, as indicated. A schematic of the GAL4-MEF2 fusions is shown at the bottom.
Figure 2
Interaction of MEF2 and HDACs 4 and 5 in yeast and mammalian cells. (A) Schematic diagrams of HDACs 4 and 5 and the different regions of the proteins encoded by cDNAs rescued as “prey” in two-hybrid screens are shown. The rescued HDAC cDNAs overlap in the 18-aa segment shown at the bottom. The HDAC catalytic domain is located at the extreme C termini of the proteins. (B and C) Coimmunoprecipitation of MEF2 factors and HDACs 4 and 5 from transfected cells. HDACs with Flag epitopes at their carboxyl termini and MEF2 factors were expressed in transiently transfected 293 T cells. Forty-eight hours after transfection, cell extracts were prepared and immunoprecipitated with anti-Flag antibody. Immunoprecipitates were then separated by SDS/PAGE and sequentially immunoblotted with anti-MEF2 or anti-Flag antibodies. (Upper) The results of anti-MEF2 Western and specific interaction of HDAC 4 and 5 with MEF2A, -C, and -D. (Lower) The results of anti-Flag (HDAC) Western blot show that comparable amounts of each HDAC were expressed in transfected cells. A schematic of the experiment is shown at the side. (C) Cell extracts were immunoprecipitated with anti-Flag antibody followed by Western blot with anti-MEF2 (Upper) or were probed by anti-MEF2 Western without prior immunoprecipitation (Lower). Deletion of the HDAC5 amino terminus prevents interaction with MEF2A, MEF2C, and MEF2D (not shown).
Figure 3
HDAC4 and -5 inhibit MEF2-dependent transcription. (A) 10T½ cells in serum-containing medium (see Materials and Methods) were transiently transfected with the MEF2-dependent reporter, 3xMEF2-luciferase, along with expression vectors for the indicated HDACs and MEF2 factors. Forty-eight hours later, cells were harvested and luciferase activity was determined. (B) 10T½ cells were transiently transfected with pG5E1b-luciferase reporter and expression vectors for GAL4-MEF2C, GAL4-MEF2C-ΔN, and the indicated HDACs and luciferase activity was determined as in A.
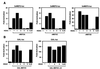
Figure 4
CaMK-dependent activation of MEF2 overcomes HDAC-mediated repression. (A) 10T½ cells in serum-containing medium were transiently transfected with the indicated expression vectors, and luciferase activity was determined. (B) 293T cells were transfected with expression plasmids encoding Flag-tagged HDAC4 with and without CaMKIV as indicated. Forty-eight hours after transfection, cell extracts were prepared, immunoprecipitated with anti-Flag antibody, and HDAC activity was determined by release of [3H]acetate from acetylated histones, as described in Materials and Methods. (C) Immunoprecipitations from extracts of cells transfected with MEF2C, HDAC5, and CaMKI expression vectors, as indicated, were performed as described in Fig. 2 B and C. In the presence of activated CaMKI, interaction between MEF2C and HDAC5 was significantly diminished. An illustration of how CaMK may activate MEF2 by dissociating HDAC is shown.
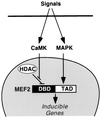
Figure 5
A model for the regulation of MEF2 activity by CaMK signaling. Hypertrophic signals that activate CaMK and MAPK lead to MEF2 activation by different mechanisms. Some stimuli, such as PE, may activate both pathways, whereas other stimuli may preferentially activate one pathway or the other. Association of HDACs 4/5 with the DNA-binding domain (DBD) of MEF2 represses MEF2 transcriptional activity. CaMK activates MEF2 by preventing association of HDACs 4/5 with MEF2. MAPK stimulates MEF2 activity by direct phosphorylation of the TAD. Together, the CaMK and MAPK pathways synergize to activate MEF2.
Similar articles
- Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes.
Nakagawa Y, Kuwahara K, Harada M, Takahashi N, Yasuno S, Adachi Y, Kawakami R, Nakanishi M, Tanimoto K, Usami S, Kinoshita H, Saito Y, Nakao K. Nakagawa Y, et al. J Mol Cell Cardiol. 2006 Dec;41(6):1010-22. doi: 10.1016/j.yjmcc.2006.08.010. Epub 2006 Oct 2. J Mol Cell Cardiol. 2006. PMID: 17011572 - CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo.
Passier R, Zeng H, Frey N, Naya FJ, Nicol RL, McKinsey TA, Overbeek P, Richardson JA, Grant SR, Olson EN. Passier R, et al. J Clin Invest. 2000 May;105(10):1395-406. doi: 10.1172/JCI8551. J Clin Invest. 2000. PMID: 10811847 Free PMC article. - Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors.
Grégoire S, Yang XJ. Grégoire S, et al. Mol Cell Biol. 2005 Mar;25(6):2273-87. doi: 10.1128/MCB.25.6.2273-2287.2005. Mol Cell Biol. 2005. PMID: 15743823 Free PMC article. - MEF2: a central regulator of diverse developmental programs.
Potthoff MJ, Olson EN. Potthoff MJ, et al. Development. 2007 Dec;134(23):4131-40. doi: 10.1242/dev.008367. Epub 2007 Oct 24. Development. 2007. PMID: 17959722 Review. - Regulatory signal transduction pathways for class IIa histone deacetylases.
Parra M, Verdin E. Parra M, et al. Curr Opin Pharmacol. 2010 Aug;10(4):454-60. doi: 10.1016/j.coph.2010.04.004. Epub 2010 May 4. Curr Opin Pharmacol. 2010. PMID: 20447866 Review.
Cited by
- Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning.
Vedantham V, Evangelista M, Huang Y, Srivastava D. Vedantham V, et al. Dev Biol. 2013 Jan 1;373(1):149-62. doi: 10.1016/j.ydbio.2012.10.017. Epub 2012 Oct 23. Dev Biol. 2013. PMID: 23085412 Free PMC article. - Acetic acid stimulates G-protein-coupled receptor GPR43 and induces intracellular calcium influx in L6 myotube cells.
Maruta H, Yamashita H. Maruta H, et al. PLoS One. 2020 Sep 30;15(9):e0239428. doi: 10.1371/journal.pone.0239428. eCollection 2020. PLoS One. 2020. PMID: 32997697 Free PMC article. - Weighted change-point method for detecting differential gene expression in breast cancer microarray data.
Wang Y, Sun G, Ji Z, Xing C, Liang Y. Wang Y, et al. PLoS One. 2012;7(1):e29860. doi: 10.1371/journal.pone.0029860. Epub 2012 Jan 20. PLoS One. 2012. PMID: 22276133 Free PMC article. - Cardiac Hypertrophy: From Pathophysiological Mechanisms to Heart Failure Development.
Caturano A, Vetrano E, Galiero R, Salvatore T, Docimo G, Epifani R, Alfano M, Sardu C, Marfella R, Rinaldi L, Sasso FC. Caturano A, et al. Rev Cardiovasc Med. 2022 May 6;23(5):165. doi: 10.31083/j.rcm2305165. eCollection 2022 May. Rev Cardiovasc Med. 2022. PMID: 39077592 Free PMC article. Review. - Apelin-APJ signaling is a critical regulator of endothelial MEF2 activation in cardiovascular development.
Kang Y, Kim J, Anderson JP, Wu J, Gleim SR, Kundu RK, McLean DL, Kim JD, Park H, Jin SW, Hwa J, Quertermous T, Chun HJ. Kang Y, et al. Circ Res. 2013 Jun 21;113(1):22-31. doi: 10.1161/CIRCRESAHA.113.301324. Epub 2013 Apr 19. Circ Res. 2013. PMID: 23603510 Free PMC article.
References
- Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. - PubMed
- Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources