A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data - PubMed (original) (raw)
. 2000 Apr;66(4):1287-97.
doi: 10.1086/302861. Epub 2000 Mar 28.
Affiliations
- PMID: 10739757
- PMCID: PMC1288195
- DOI: 10.1086/302861
A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data
J A Douglas et al. Am J Hum Genet. 2000 Apr.
Abstract
The identification of genes contributing to complex diseases and quantitative traits requires genetic data of high fidelity, because undetected errors and mutations can profoundly affect linkage information. The recent emphasis on the use of the sibling-pair design eliminates or decreases the likelihood of detection of genotyping errors and marker mutations through apparent Mendelian incompatibilities or close double recombinants. In this article, we describe a hidden Markov method for detecting genotyping errors and mutations in multilocus linkage data. Specifically, we calculate the posterior probability of genotyping error or mutation for each sibling-pair-marker combination, conditional on all marker data and an assumed genotype-error rate. The method is designed for use with sibling-pair data when parental genotypes are unavailable. Through Monte Carlo simulation, we explore the effects of map density, marker-allele frequencies, marker position, and genotype-error rate on the accuracy of our error-detection method. In addition, we examine the impact of genotyping errors and error detection and correction on multipoint linkage information. We illustrate that even moderate error rates can result in substantial loss of linkage information, given efforts to fine-map a putative disease locus. Although simulations suggest that our method detects </=50% of genotyping errors, it generally flags those errors that have the largest impact on linkage results. For high-resolution genetic maps, removal of the errors identified by our method restores most or nearly all the lost linkage information and can be accomplished without generating false evidence for linkage by removing incorrectly identified errors.
Figures
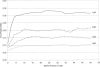
Figure 1
True-positive rate versus false-positive rate, assuming a genotype-error rate of .01, markers with four equally frequent alleles, and a 100-cM map with markers equally spaced at 1-, 2-, 3-, or 5-cM intervals. True-positive rates were based on introduction of random genotype error for at least one member of the sibling pair at the marker in the middle of the map.
Figure 2
True-positive rate as a function of marker distance from the end of the map, on the assumption of a true genotype-error rate of .01, markers with four equally frequent alleles, and a 100-cM map with markers equally spaced at 1-, 2-, 3-, or 5-cM intervals. True-positive rates were based on introduction of random genotype error for at least one member of the sibling pair at the positioned marker. The false-positive rate was fixed at .001.
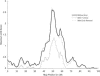
Figure 3
Impact of genotyping errors and error detection on linkage information, as measured by maximum LOD score. Linkage data were simulated for 400 sibling pairs on the assumption of a recurrence-risk ratio
λ=1.25
. Results shown are based on a 100-cM map, markers with four equally frequent alleles equally spaced at 1-cM intervals, and a disease locus placed at 50.5 cM. Random genotype errors were introduced in each sibling at each marker with probability equal to the genotype-error rate of 1%. Error removal (— —) gives linkage results after removing sibling-pair-marker combinations with high posterior error probabilities, on the assumption of a false-positive rate of .001.
Similar articles
- Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data.
Douglas JA, Skol AD, Boehnke M. Douglas JA, et al. Am J Hum Genet. 2002 Feb;70(2):487-95. doi: 10.1086/338919. Epub 2002 Jan 8. Am J Hum Genet. 2002. PMID: 11791214 Free PMC article. - Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters.
Göring HH, Terwilliger JD. Göring HH, et al. Am J Hum Genet. 2000 Apr;66(4):1298-309. doi: 10.1086/302846. Epub 2000 Mar 23. Am J Hum Genet. 2000. PMID: 10731467 Free PMC article. - The impact of genotyping error on family-based analysis of quantitative traits.
Abecasis GR, Cherny SS, Cardon LR. Abecasis GR, et al. Eur J Hum Genet. 2001 Feb;9(2):130-4. doi: 10.1038/sj.ejhg.5200594. Eur J Hum Genet. 2001. PMID: 11313746 - Linkage analysis of genetic disorders.
Taylor EW, Xu J, Jabs EW, Meyers DA. Taylor EW, et al. Methods Mol Biol. 1997;68:11-25. doi: 10.1385/0-89603-482-8:11. Methods Mol Biol. 1997. PMID: 9055247 Review. - Recent developments in genomewide association scans: a workshop summary and review.
Thomas DC, Haile RW, Duggan D. Thomas DC, et al. Am J Hum Genet. 2005 Sep;77(3):337-45. doi: 10.1086/432962. Epub 2005 Aug 1. Am J Hum Genet. 2005. PMID: 16080110 Free PMC article. Review.
Cited by
- A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q.
Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, Frayling TM, Bell JI, Lathrop GM, Bennett A, Dhillon R, Fletcher C, Groves CJ, Jones E, Prestwich P, Simecek N, Rao PV, Wishart M, Bottazzo GF, Foxon R, Howell S, Smedley D, Cardon LR, Menzel S, McCarthy MI. Wiltshire S, et al. Am J Hum Genet. 2001 Sep;69(3):553-69. doi: 10.1086/323249. Epub 2001 Aug 1. Am J Hum Genet. 2001. PMID: 11484155 Free PMC article. - Genotyping error detection through tightly linked markers.
Zou G, Pan D, Zhao H. Zou G, et al. Genetics. 2003 Jul;164(3):1161-73. doi: 10.1093/genetics/164.3.1161. Genetics. 2003. PMID: 12871922 Free PMC article. - Genetic mapping in the presence of genotyping errors.
Cartwright DA, Troggio M, Velasco R, Gutin A. Cartwright DA, et al. Genetics. 2007 Aug;176(4):2521-7. doi: 10.1534/genetics.106.063982. Epub 2007 Feb 4. Genetics. 2007. PMID: 17277374 Free PMC article. - Minimum-recombinant haplotyping in pedigrees.
Qian D, Beckmann L. Qian D, et al. Am J Hum Genet. 2002 Jun;70(6):1434-45. doi: 10.1086/340610. Epub 2002 Apr 25. Am J Hum Genet. 2002. PMID: 11992251 Free PMC article. - Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps.
Evans DM, Cardon LR. Evans DM, et al. Am J Hum Genet. 2004 Oct;75(4):687-92. doi: 10.1086/424696. Epub 2004 Aug 13. Am J Hum Genet. 2004. PMID: 15311375 Free PMC article.
References
Electronic-Database Information
- ASPEX (Affected Sib Pairs EXclusion Map) version 2.2, ftp://lahmed.stanford.edu/pub/aspex/index.html
- SIBMED program, <http://www. sph.umich.edu/group/statgen/software>
References
- Baum LE (1972) An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes. Inequalities 3:11–18
- Dracopoli NC, O’Connell P, Elsner TI, Lalouel J-M, White RL, Buetow K, Nishimura DY, et al (1991) The CEPH consortium linkage map of human chromosome 1. Genomics 9:686–700 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HG00040/HG/NHGRI NIH HHS/United States
- R01 HG00376/HG/NHGRI NIH HHS/United States
- R01 HG000376/HG/NHGRI NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- MH59490/MH/NIMH NIH HHS/United States
- R01 MH059490/MH/NIMH NIH HHS/United States
- R37 MH059490/MH/NIMH NIH HHS/United States
- R01 GM053275/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases