SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition - PubMed (original) (raw)
SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition
E E Patton et al. EMBO J. 2000.
Abstract
Progression through the cell cycle requires the coordination of basal metabolism with the cell cycle and growth machinery. Repression of the sulfur gene network is mediated by the ubiquitin ligase SCF(Met30), which targets the transcription factor Met4p for degradation. Met30p is an essential protein in yeast. We have found that a met4Deltamet30Delta double mutant is viable, suggesting that the essential function of Met30p is to control Met4p. In support of this hypothesis, a Met4p mutant unable to activate transcription does not cause inviability in a met30Delta strain. Also, overexpression of an unregulated Met4p mutant is lethal in wild-type cells. Under non-permissive conditions, conditional met30Delta strains arrest as large, unbudded cells with 1N DNA content, at or shortly after the pheromone arrest point. met30Delta conditional mutants fail to accumulate CLN1 and CLN2, but not CLN3 mRNAs, even when CLN1 and CLN2 are expressed from strong heterologous promoters. One or more genes under the regulation of Met4p may delay the progression from G(1) into S phase through specific regulation of critical G(1) phase mRNAs.
Figures
Fig. 1. (A) Simplified view of sulfur amino acid biosynthesis in yeast. (B) The met4Δ disruption mutation specifically suppresses the lethality induced by the loss-of-function mutation met30Δ. See Materials and methods for details of the crosses.
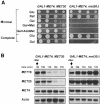
Fig. 2. Expression of Met4p from the GAL1 promoter is lethal in met30Δ cells. (A) Serial dilutions of the CC932-6D (met4::GAL1–MET4 MET30) and CC932-8B (met4::GAL1–MET4 met30::LEU2) strains were plated on media containing 2% glucose (Glu), raffinose (Raf) or galactose (Gal) as carbon source in the absence or presence of 1 mM
l
-methionine (Met) or 0.2 mM AdoMet. (B) CC932-6D (met4::GAL1–MET4 MET30) and CC932-8B (met4::GAL1–MET4 met30::LEU2) cells were grown in raffinose medium and expression was induced by transferring the cells to a fresh galactose (2%) medium for 90 min. A repressing amount of
l
-Met (1 mM) was then added and total RNA was extracted at the times indicated. Expression of MET16, MET25 and GAL1–MET4 were determined by Northern analysis. The actin probe was used as a control of the amount of RNA loaded.
Fig. 3. (A) Expression of Met4p derivatives in met4Δ met30Δ cells. Schematic representations of the modified Met4p derivatives expressed from the GAL1 promoter region are shown. Plasmids encoding the fusion genes were introduced into CC807-1C (met4::TRP1 met30::LEU2) cells. Serial dilutions of the resulting transformants were plated on media containing 2% raffinose or galactose as carbon source. (B) Expression of Met4p derivatives in met4Δ MET30 cells. Details of the deletions are scaled up. Plasmids encoding the fusion genes were introduced into CD106 (met4::TRP1) cells. Serial dilutions of the resulting transformants were plated on media containing 2% raffinose or galactose as carbon source.
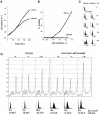
Fig. 4. Conditional met30Δ mutant cells arrest as large unbudded cells with 1N DNA. (A) CC932-6D (met4::GAL1–MET4 MET30) (▪) and CC932-8B (met4::GAL1–MET4 met30::LEU2) (•) cells were grown in the presence of 2% raffinose as carbon source. At the time indicated by the arrow, 2% galactose was added to each culture and growth was followed by measuring the OD650 for the times indicated. (B) Wild type, met4::GAL1–MET4 MET30 and met4::GAL1–MET4 met30::LEU2 were grown in the presence of raffinose, cultures were divided into two, filtered and transferred to raffinose or galactose medium. FACS analyses were then performed at the times indicated. (C) CC807-1C (met4::TRP1 met30::LEU2) cells were transformed by the pGal316, pGalMet4-1 or pGalMet4Δ12 plasmids. Resulting transformants were grown in the presence of 2% raffinose as carbon source. Cultures were then filtered and transferred to a galactose (2%) medium. Photographs of the cells were then taken at the times indicated. (D) Budding index of the cells grown in (A). (E) CC807-1C (met4::TRP1 met30::LEU2) or CD106 (met4::TRP1) were transformed by the plasmids indicated. Resulting transformants were grown in raffinose medium to early log phase and arrested by α factor for 3 h in 2% galactose. The cells were released from α factor by washing with fresh galactose medium and analyzed for their DNA content by flow cytometry at the times indicated.
Fig. 5. The essential function of MET30 is required at or after the pheromone arrest position, but prior to the initiation of budding or DNA replication. (A) Wild-type, met4::GAL1–MET4 and met4::GAL1–MET4 met30Δ cells were grown in rich raffinose medium to early log phase and arrested with α factor for 2 h. The cells were then split into two cultures and raffinose added to one, while galactose was added to another for 20 min. The cells were released from α factor by washing with fresh medium and time points taken at the designated intervals. (B) Photographs of the cells in (A) demonstrate that the met4::GAL1–MET4 met30Δ arrest as large unbudded cells. The tear shape of the cells is due to effects of the pheromone. (C) MET30 is required before the cdc7-1 arrest point. Wild-type, cdc7-1 and cdc7-1 met4::GAL1–MET4 met30Δ cultures were incubated at 37°C for 2 h, had galactose added for 30 min, and were then shifted to 25°C and samples taken at the time indicated.
Fig. 6. Conditional G1-arrested met30Δ mutants are slowed for cell growth and translation. Cultures of small G1 met4::GAL1–MET4 met30Δ cells grown in raffinose were collected by centrifugal elutriation, divided into two, and raffinose added to one culture and galactose to the other. Samples were taken at the times indicated to measure (A) cell volume, (B) budding index and (C) DNA content by flow cytometry. (D) Polysome profiles of wild-type or met4::GAL1–MET4 met30Δ cells through the Start position. G1 cells were collected by centrifugal elutriation in a rich, raffinose medium, galactose was added at time 0, and samples were taken at the times indicated for polysome profiles, flow cytometry and budding analysis.
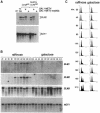
Fig. 7. Conditional met30Δ mutants lack CLN1 and CLN2 mRNA transcripts. (A) Northern analysis of CLN mRNA transcripts demonstrates a lack of CLN2 mRNA expressed from the endogenous or the ADH1 promoter in galactose-grown met4::GAL1–MET4 met30Δ cells. (B) CLN1 and CLN2, but not CLN3 mRNAs, fail to accumulate in elutriated G1 met4::GAL1–MET4 met30Δ cells grown in galactose, but accumulate normally in met4::GAL1–MET4 met30Δ cells grown in raffinose. Cells were grown in a rich, raffinose medium, the culture split into two, and raffinose added to one culture and galactose to the other. Samples were taken for (B) Northern analysis and (C) flow cytometry.
Similar articles
- Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30 )complex.
Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Rouillon A, et al. EMBO J. 2000 Jan 17;19(2):282-94. doi: 10.1093/emboj/19.2.282. EMBO J. 2000. PMID: 10637232 Free PMC article. - Identification of residues in the WD-40 repeat motif of the F-box protein Met30p required for interaction with its substrate Met4p.
Brunson LE, Dixon C, LeFebvre A, Sun L, Mathias N. Brunson LE, et al. Mol Genet Genomics. 2005 Jun;273(5):361-70. doi: 10.1007/s00438-005-1137-6. Epub 2005 May 10. Mol Genet Genomics. 2005. PMID: 15883825 - Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4.
Kaiser P, Flick K, Wittenberg C, Reed SI. Kaiser P, et al. Cell. 2000 Aug 4;102(3):303-14. doi: 10.1016/s0092-8674(00)00036-2. Cell. 2000. PMID: 10975521 - The F-box protein Met30 is required for multiple steps in the budding yeast cell cycle.
Su NY, Flick K, Kaiser P. Su NY, et al. Mol Cell Biol. 2005 May;25(10):3875-85. doi: 10.1128/MCB.25.10.3875-3885.2005. Mol Cell Biol. 2005. PMID: 15870262 Free PMC article. - Metabolism of sulfur amino acids in Saccharomyces cerevisiae.
Thomas D, Surdin-Kerjan Y. Thomas D, et al. Microbiol Mol Biol Rev. 1997 Dec;61(4):503-32. doi: 10.1128/mmbr.61.4.503-532.1997. Microbiol Mol Biol Rev. 1997. PMID: 9409150 Free PMC article. Review.
Cited by
- Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii.
Barbosa C, Mendes-Faia A, Lage P, Mira NP, Mendes-Ferreira A. Barbosa C, et al. Microb Cell Fact. 2015 Aug 28;14:124. doi: 10.1186/s12934-015-0318-1. Microb Cell Fact. 2015. PMID: 26314747 Free PMC article. - Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase.
Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Aghajan M, et al. Nat Biotechnol. 2010 Jul;28(7):738-42. doi: 10.1038/nbt.1645. Epub 2010 Jun 27. Nat Biotechnol. 2010. PMID: 20581845 Free PMC article. - Determinants of Swe1p degradation in Saccharomyces cerevisiae.
McMillan JN, Theesfeld CL, Harrison JC, Bardes ES, Lew DJ. McMillan JN, et al. Mol Biol Cell. 2002 Oct;13(10):3560-75. doi: 10.1091/mbc.e02-05-0283. Mol Biol Cell. 2002. PMID: 12388757 Free PMC article. - The Aspergillus nidulans swoC1 mutant shows defects in growth and development.
Lin X, Momany M. Lin X, et al. Genetics. 2003 Oct;165(2):543-54. doi: 10.1093/genetics/165.2.543. Genetics. 2003. PMID: 14573468 Free PMC article. - Genetic analysis of B55alpha/Cdc55 protein phosphatase 2A subunits: association with the adenovirus E4orf4 protein.
Zhang Z, Mui MZ, Chan F, Roopchand DE, Marcellus RC, Blanchette P, Li S, Berghuis AM, Branton PE. Zhang Z, et al. J Virol. 2011 Jan;85(1):286-95. doi: 10.1128/JVI.01381-10. Epub 2010 Nov 3. J Virol. 2011. PMID: 21047956 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases