A Sm-like protein complex that participates in mRNA degradation - PubMed (original) (raw)
A Sm-like protein complex that participates in mRNA degradation
E Bouveret et al. EMBO J. 2000.
Abstract
In eukaryotes, seven Sm proteins bind to the U1, U2, U4 and U5 spliceosomal snRNAs while seven Smlike proteins (Lsm2p-Lsm8p) are associated with U6 snRNA. Another yeast Sm-like protein, Lsm1p, does not interact with U6 snRNA. Surprisingly, using the tandem affinity purification (TAP) method, we identified Lsm1p among the subunits associated with Lsm3p. Coprecipitation experiments demonstrated that Lsm1p, together with Lsm2p-Lsm7p, forms a new seven-subunit complex. We purified the two related Sm-like protein complexes and identified the proteins recovered in the purified preparations by mass spectrometry. This confirmed the association of the Lsm2p-Lsm8p complex with U6 snRNA. In contrast, the Lsm1p-Lsm7p complex is associated with Pat1p and Xrn1p exoribonuclease, suggesting a role in mRNA degradation. Deletions of LSM1, 6, 7 and PAT1 genes increased the half-life of reporter mRNAs. Interestingly, accumulating mRNAs were capped, suggesting a block in mRNA decay at the decapping step. These results indicate the involvement of a new conserved Sm-like protein complex and a new factor, Pat1p, in mRNA degradation and suggest a physical connection between decapping and exonuclease trimming.
Figures
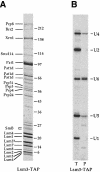
Fig. 1. Purification of the Lsm3p-interacting proteins. From 2 l of culture of Lsm3p–TAP-expressing strain, the Lsm3p-interacting proteins were purified by the TAP method (Rigaut et al., 1999). (A) The purified material was fractionated on a 7–25% gradient SDS gel, which was then Coomassie Blue stained. Proteins identified either by MALDI or by nano-electrospray tandem mass spectrometry in this purification and/or similar purifications are indicated on the left. Some faint bands that were not reproducibly found in different purifications are not labeled. Molecular weight markers are indicated on the right. An asterisk after Lsm3 (Lsm3*) indicates that it still carries a part of the TAP tag. Pat1d stands for putative degradation products of Pat1. SmB was identified from a piece of gel containing the closely spaced bands indicated. (B) Total RNAs were recovered from the extract before purification (T) and from the purified fraction (P) and their U snRNAs content analyzed by primer extension. Signals corresponding to the different U snRNAs are indicated on the right. There is a 5–fold excess loaded for the purified fraction compared with the total RNAs.
Fig. 2. Lsm1p is in a complex with the Sm-like proteins Lsm2p–Lsm7p. Extracts were prepared from strains expressing a CBP-tagged Lsm1 protein (even lanes) or wild-type Lsm1p (odd lanes) in addition to ProtA-tagged Sm-like proteins (see Materials and methods). After precipitation on calmodulin beads, the presence of coprecipitated ProtA-tagged Sm-like proteins with the Lsm1p–CBP fusion was assayed by 15% SDS–PAGE and Western blotting. Proteins present in the supernatants and in the pellets are shown. There is an 8–fold excess of pellet loaded compared with the supernatants.
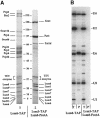
Fig. 3. Purifications of the Lsm1p- and Lsm8p-containing complexes. Extracts from strains expressing either Lsm8p–TAP or Lsm3–TAP/Lsm8–ProtA fusions were prepared from cultures of 4 l. The complexes were purified using the TAP method (Rigaut et al., 1999). (A) Purified fractions were fractionated on a 7–25% gradient SDS gel. The figure shows the Coomassie Blue staining of this gel. The names of the proteins, which were identified either by MALDI or by nano-electrospray tandem mass spectrometry, are indicated on the sides. The molecular weight markers are indicated in the middle. The asterisk in Lsm8* and Lsm3* indicates that these proteins still carry part of the TAP tag. Pat1d stands for a putative degradation product of Pat1. Contaminants coming from the TEV preparation are indicated. (B) RNAs extracted from the extracts and the purifications were analyzed by primer extension for their U snRNA content. (T) Total RNAs in the extract before purification, (P) RNAs in the purified fraction. The different U snRNAs are indicated on the right. There is a 5–fold excess loaded for the purification fraction compared with the extract fraction. (C) Identification of Lsm5p in the Lsm1p-containing complex (A, lane 2) with tandem mass spectrometry. Two peptides of the protein were identified by comparison of the spectrum with a blank to distinguish them from autolysis products of the enzyme and common keratin peptides. Both peptides were fragmented and allowed independently the identification of Lsm5p (SwissProt P40089, hypothetical 10.4 kDa protein). One of the peptides identified and the corresponding spectra are shown.
Fig. 3. Purifications of the Lsm1p- and Lsm8p-containing complexes. Extracts from strains expressing either Lsm8p–TAP or Lsm3–TAP/Lsm8–ProtA fusions were prepared from cultures of 4 l. The complexes were purified using the TAP method (Rigaut et al., 1999). (A) Purified fractions were fractionated on a 7–25% gradient SDS gel. The figure shows the Coomassie Blue staining of this gel. The names of the proteins, which were identified either by MALDI or by nano-electrospray tandem mass spectrometry, are indicated on the sides. The molecular weight markers are indicated in the middle. The asterisk in Lsm8* and Lsm3* indicates that these proteins still carry part of the TAP tag. Pat1d stands for a putative degradation product of Pat1. Contaminants coming from the TEV preparation are indicated. (B) RNAs extracted from the extracts and the purifications were analyzed by primer extension for their U snRNA content. (T) Total RNAs in the extract before purification, (P) RNAs in the purified fraction. The different U snRNAs are indicated on the right. There is a 5–fold excess loaded for the purification fraction compared with the extract fraction. (C) Identification of Lsm5p in the Lsm1p-containing complex (A, lane 2) with tandem mass spectrometry. Two peptides of the protein were identified by comparison of the spectrum with a blank to distinguish them from autolysis products of the enzyme and common keratin peptides. Both peptides were fragmented and allowed independently the identification of Lsm5p (SwissProt P40089, hypothetical 10.4 kDa protein). One of the peptides identified and the corresponding spectra are shown.
Fig. 4. Pat1p is in complex with the Sm-like proteins Lsm1p–Lsm7p. Extracts were prepared from strains expressing a TAP-tagged Pat1 protein together with ProtA-tagged Sm-like proteins (see Materials and methods). After precipitation of Pat1p–TAP on calmodulin beads, the presence of a coprecipitated ProtA-tagged Sm-like protein was assayed by 15% SDS–PAGE and Western blotting. Proteins present in the supernatants and the pellets are shown. There is an 8–fold excess of pellet loaded compared with the supernatant.
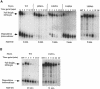
Fig. 5. MFA2pG mRNA is stabilized in lsm1, 6, 7 and pat1 null mutants. Wild-type lsm1, 5, 6, 7 and pat1 null mutant yeast strains carrying the GAL1:MFA2pG reporter (RP485; Decker and Parker, 1993) were grown in 2% galactose-containing minimal medium to an OD600 of 0.4. Transcription was then repressed with 2% glucose at time 0. At the indicated time-points, cells were harvested, RNAs extracted and then analyzed on a 6% denaturing acrylamide gel and by Northern blotting using the oligo(C) probe bo29. As an internal loading standard, we used the oligonucleotide bo36 to detect the scR1 transcript (not shown). For the _lsm6_Δ, _pat1_Δ and wild-type strains, the samples from zero time-point were deadenylated (0dT) to serve as size marker for deadenylated species. The calculated half-life of the MFA2pG reporter in each mutant is indicated below the gels. (A) Wild-type, _lsm1_Δ, _lsm6_Δ, _lsm7_Δ and _lsm5_Δ strains grown at 37°C. (B) Wild-type and _pat1_Δ strains grown at 30°C.
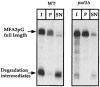
Fig. 6. mRNAs that accumulate in _pat1_Δ mutant are capped. RNAs from time-points 0 min for the wild-type strain and 6 min for the _pat1_Δ strain corresponding to Figure 5 were used. These RNAs were immunoprecipitated with an antiserum directed against the 7–methyl cap structure as described in Materials and methods and then analyzed by Northern blotting as described in Figure 5. I, input; P, pellet; SN, supernatant. The relative amounts loaded for each fraction are identical.
Fig. 7. The pat1 null mutant is not affected in PGK1NSpG mRNA degradation. The wild-type and _pat1_Δ strains carrying the GAL1:PGK1NSpG reporter (RP611; Muhlrad and Parker, 1994) were grown in 2% galactose-containing minimal medium to an OD600 of 0.4 (lanes labeled ss for steady state). Transcription was then repressed with 2% glucose at time 0. At the indicated time-points, cells were harvested, RNAs extracted and then analyzed on a 1% formaldehyde–agarose gel and by Northern blotting using the oligo(C) probe bo29. As an internal loading standard, we used the oligonucleotide bo36 to detect the scR1 transcript.
Fig. 8. Composition of the two Lsm complexes in yeast. Lsm protein organization characterized in the present study is summarized. Names of proteins are indicated, except for Lsm proteins, which are only indicated by their number. The order of Lsm proteins in the putative heptameric rings is based on similarity between Sm and Sm-like proteins (Salgado-Garrido et al., 1999) and the model proposed for Sm proteins by Kambach et al. (1999). Interactions of non Sm-like proteins with the Sm-like complexes may be direct or indirect and are not known precisely (see Discussion). Prp24p is not a component of U4/U6⋅U5 tri-snRNP and therefore is shown separately.
Similar articles
- Yeast Sm-like proteins function in mRNA decapping and decay.
Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Tharun S, et al. Nature. 2000 Mar 30;404(6777):515-8. doi: 10.1038/35006676. Nature. 2000. PMID: 10761922 - Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins.
Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, Legrain P. Fromont-Racine M, et al. Yeast. 2000 Jun 30;17(2):95-110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. Yeast. 2000. PMID: 10900456 Free PMC article. - Purification and analysis of the decapping activator Lsm1p-7p-Pat1p complex from yeast.
Tharun S. Tharun S. Methods Enzymol. 2008;448:41-55. doi: 10.1016/S0076-6879(08)02603-7. Methods Enzymol. 2008. PMID: 19111170 - Functions of Lsm proteins in mRNA degradation and splicing.
He W, Parker R. He W, et al. Curr Opin Cell Biol. 2000 Jun;12(3):346-50. doi: 10.1016/s0955-0674(00)00098-3. Curr Opin Cell Biol. 2000. PMID: 10801455 Review. - Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.
Tucker M, Parker R. Tucker M, et al. Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
- The TUTase URT1 connects decapping activators and prevents the accumulation of excessively deadenylated mRNAs to avoid siRNA biogenesis.
Scheer H, de Almeida C, Ferrier E, Simonnot Q, Poirier L, Pflieger D, Sement FM, Koechler S, Piermaria C, Krawczyk P, Mroczek S, Chicher J, Kuhn L, Dziembowski A, Hammann P, Zuber H, Gagliardi D. Scheer H, et al. Nat Commun. 2021 Feb 26;12(1):1298. doi: 10.1038/s41467-021-21382-2. Nat Commun. 2021. PMID: 33637717 Free PMC article. - Eukaryotic mRNA decapping factors: molecular mechanisms and activity.
He F, Jacobson A. He F, et al. FEBS J. 2023 Nov;290(21):5057-5085. doi: 10.1111/febs.16626. Epub 2022 Sep 30. FEBS J. 2023. PMID: 36098474 Free PMC article. Review. - The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex.
Vilela C, Velasco C, Ptushkina M, McCarthy JE. Vilela C, et al. EMBO J. 2000 Aug 15;19(16):4372-82. doi: 10.1093/emboj/19.16.4372. EMBO J. 2000. PMID: 10944120 Free PMC article. - A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting.
Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. Tritschler F, et al. Mol Cell Biol. 2007 Dec;27(24):8600-11. doi: 10.1128/MCB.01506-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923697 Free PMC article. - Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly.
Tardiff DF, Rosbash M. Tardiff DF, et al. RNA. 2006 Jun;12(6):968-79. doi: 10.1261/rna.50506. Epub 2006 Apr 17. RNA. 2006. PMID: 16618970 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous