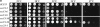Structure and assembly of the Nup84p complex - PubMed (original) (raw)
Structure and assembly of the Nup84p complex
S Siniossoglou et al. J Cell Biol. 2000.
Abstract
The Nup84p complex consists of five nucleoporins (Nup84p, Nup85p, Nup120p, Nup145p-C, and Seh1p) and Sec13p, a bona fide subunit of the COPII coat complex. We show that a pool of green fluorescent protein-tagged Sec13p localizes to the nuclear pores in vivo, and identify sec13 mutant alleles that are synthetically lethal with nup85Delta and affect the localization of a green fluorescent protein-Nup49p reporter protein. In the electron microscope, sec13 mutants exhibit structural defects in nuclear pore complex (NPC) and nuclear envelope organization. For the assembly of the complex, Nup85p, Nup120p, and Nup145p-C are essential. A highly purified Nup84p complex was isolated from yeast under native conditions and its molecular mass was determined to be 375 kD by quantitative scanning transmission electron microscopy and analytical ultracentrifugation, consistent with a monomeric complex. Furthermore, the Nup84p complex exhibits a Y-shaped, triskelion-like morphology 25 nm in diameter in the transmission electron microscope. Thus, the Nup84p complex constitutes a paradigm of an NPC structural module with distinct composition, structure, and a role in nuclear mRNA export and NPC bio- genesis.
Figures
Figure 1
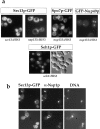
A pool of Sec13p-GFP localizes to the nuclear pores. (a) The sec13 − /nup133 − double mutant complemented by either SEC13-GFP and NUP133 (sec13::HIS3) or by SEC13-GFP alone (nup133::HIS3) was analyzed by fluorescence microscopy (upper panel). In addition, Spo7p-GFP (Siniossoglou et al. 1998) and GFP-Nup49p were tested in the nup133 − cells (upper panel). For comparison, the seh1::HIS3 disruption mutant complemented by plasmid-borne SEH1-GFP was analyzed in the fluorescence microscope (lower panel). (b) The sec13 − /nup133 − double mutant expressing Sec13p-GFP was analyzed by indirect immunofluorescence microscopy using a monoclonal anti-Nsp1p antibody. It was also stained for DNA.
Figure 2
Novel sec13 thermosensitive alleles that show defects in GFP-Nup49p location and nuclear membrane morphology. (a) Growth properties of novel sec13 thermosensitive mutants. Precultures derived from the sec13::HIS3 disrupted strain transformed with ARS/CEN plasmids containing the indicated SEC13 wild-type and sec13 ts alleles were diluted in growth medium and equivalent amounts of cells (diluted in 10_−_ steps) were spotted onto yeast extract–peptone–d-glucose plates. All SEC13 and sec13 alleles are tagged with one IgG-binding domain from ProtA in their COOH termini. For control, the sec23-1 ts mutant was also included in this analysis. It was grown for 3 d at the indicated temperatures. (b) Location of GFP-Nup49p and GFP-Pus1p in SEC13 + and sec13 and sec23-1 ts mutants as determined by fluorescence microscopy. Cells were grown at 23°C and shifted for 5 h to either 32 or 35°C. (c) Thin-section EM of SEC13, sec13-3, sec13-14, and sec13-34 cells, shifted for 5 h to 35°C. Filled arrowheads point to normal nuclear pores, arrows to nuclear envelope herniations, and the open arrow to a long nuclear membrane extension with a herniation morphology at the tip. Bars, 0.5 μm.
Figure 2
Novel sec13 thermosensitive alleles that show defects in GFP-Nup49p location and nuclear membrane morphology. (a) Growth properties of novel sec13 thermosensitive mutants. Precultures derived from the sec13::HIS3 disrupted strain transformed with ARS/CEN plasmids containing the indicated SEC13 wild-type and sec13 ts alleles were diluted in growth medium and equivalent amounts of cells (diluted in 10_−_ steps) were spotted onto yeast extract–peptone–d-glucose plates. All SEC13 and sec13 alleles are tagged with one IgG-binding domain from ProtA in their COOH termini. For control, the sec23-1 ts mutant was also included in this analysis. It was grown for 3 d at the indicated temperatures. (b) Location of GFP-Nup49p and GFP-Pus1p in SEC13 + and sec13 and sec23-1 ts mutants as determined by fluorescence microscopy. Cells were grown at 23°C and shifted for 5 h to either 32 or 35°C. (c) Thin-section EM of SEC13, sec13-3, sec13-14, and sec13-34 cells, shifted for 5 h to 35°C. Filled arrowheads point to normal nuclear pores, arrows to nuclear envelope herniations, and the open arrow to a long nuclear membrane extension with a herniation morphology at the tip. Bars, 0.5 μm.
Figure 2
Novel sec13 thermosensitive alleles that show defects in GFP-Nup49p location and nuclear membrane morphology. (a) Growth properties of novel sec13 thermosensitive mutants. Precultures derived from the sec13::HIS3 disrupted strain transformed with ARS/CEN plasmids containing the indicated SEC13 wild-type and sec13 ts alleles were diluted in growth medium and equivalent amounts of cells (diluted in 10_−_ steps) were spotted onto yeast extract–peptone–d-glucose plates. All SEC13 and sec13 alleles are tagged with one IgG-binding domain from ProtA in their COOH termini. For control, the sec23-1 ts mutant was also included in this analysis. It was grown for 3 d at the indicated temperatures. (b) Location of GFP-Nup49p and GFP-Pus1p in SEC13 + and sec13 and sec23-1 ts mutants as determined by fluorescence microscopy. Cells were grown at 23°C and shifted for 5 h to either 32 or 35°C. (c) Thin-section EM of SEC13, sec13-3, sec13-14, and sec13-34 cells, shifted for 5 h to 35°C. Filled arrowheads point to normal nuclear pores, arrows to nuclear envelope herniations, and the open arrow to a long nuclear membrane extension with a herniation morphology at the tip. Bars, 0.5 μm.
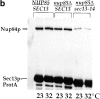
Figure 3
Genetic interaction between the sec13-34 and _nup85_Δ mutant alleles. (a) Growth curves of the sec13-34/nup85Δ double mutant and of the corresponding sec13-34 and nup85Δ single mutants at 23 and 32°C. The nup85Δ allele is derived from a partial gene disruption and expresses a Nup85p protein which is NH2-terminally truncated (Siniossoglou et al. 1996). (b) Affinity-purification of Sec13p-ProtA from NUP85/SEC13, nup85Δ/SEC13, and nup85Δ/sec13-14 strains grown at 23°C or shifted for 12 h to 32°C. The purified wild-type or mutant Sec13p-ProtA fusion proteins were analyzed by Western blotting using anti-Nup84p antibodies. Indicated are the positions of Sec13p-ProtA and Nup84p.
Figure 3
Genetic interaction between the sec13-34 and _nup85_Δ mutant alleles. (a) Growth curves of the sec13-34/nup85Δ double mutant and of the corresponding sec13-34 and nup85Δ single mutants at 23 and 32°C. The nup85Δ allele is derived from a partial gene disruption and expresses a Nup85p protein which is NH2-terminally truncated (Siniossoglou et al. 1996). (b) Affinity-purification of Sec13p-ProtA from NUP85/SEC13, nup85Δ/SEC13, and nup85Δ/sec13-14 strains grown at 23°C or shifted for 12 h to 32°C. The purified wild-type or mutant Sec13p-ProtA fusion proteins were analyzed by Western blotting using anti-Nup84p antibodies. Indicated are the positions of Sec13p-ProtA and Nup84p.
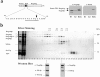
Figure 4
Distinct domains within Nup85p are required for interaction with the various members of the Nup84p complex. (a) Growth of nup85ΔC and nup85ΔN strains at 23, 30, and 35°C. A nup85::HIS3 null deletion mutant was transformed with the two nup85 truncation constructs and the corresponding strains were spotted onto yeast extract–peptone–d-glucose plates at the indicated temperatures. (b) Affinity-purification ProtA–tagged Nup85p full-length (FL) and of the Nup85pΔN (Δ1–182) and Nup85pΔC (Δ453–745) constructs. Shown is a Coomassie-stained gel of the purified complexes (the fusion proteins are indicated by asterisks, and the components of the Nup84p complex by lines), and a Western blot (right panel, only the relevant area from each blot is shown) using anti–ProtA, anti-Nup84p, anti-Nup145Cp, anti-Seh1p, and anti-Sec13p antibodies. (c) Deletion of the Nup85p COOH-terminal domain causes NPC clustering. The corresponding strains were transformed with a plasmid expressing GFP-Nup49p, shifted for 3 h to 37°C and the distribution of this NPC reporter was analyzed in the fluorescence microscope. (d) Both the COOH and NH2 domains of Nup85p are required for efficient nuclear mRNA export. The nup85ΔC and nup85ΔN mutants as well as a wild-type control (NUP85 +) were grown for 3 h at 37°C, fixed, and hybridized with a oligo(dT)-FITC probe to reveal poly(A)+ RNA distribution. Cells were also stained with Hoechst 33258 (DNA).
Figure 4
Distinct domains within Nup85p are required for interaction with the various members of the Nup84p complex. (a) Growth of nup85ΔC and nup85ΔN strains at 23, 30, and 35°C. A nup85::HIS3 null deletion mutant was transformed with the two nup85 truncation constructs and the corresponding strains were spotted onto yeast extract–peptone–d-glucose plates at the indicated temperatures. (b) Affinity-purification ProtA–tagged Nup85p full-length (FL) and of the Nup85pΔN (Δ1–182) and Nup85pΔC (Δ453–745) constructs. Shown is a Coomassie-stained gel of the purified complexes (the fusion proteins are indicated by asterisks, and the components of the Nup84p complex by lines), and a Western blot (right panel, only the relevant area from each blot is shown) using anti–ProtA, anti-Nup84p, anti-Nup145Cp, anti-Seh1p, and anti-Sec13p antibodies. (c) Deletion of the Nup85p COOH-terminal domain causes NPC clustering. The corresponding strains were transformed with a plasmid expressing GFP-Nup49p, shifted for 3 h to 37°C and the distribution of this NPC reporter was analyzed in the fluorescence microscope. (d) Both the COOH and NH2 domains of Nup85p are required for efficient nuclear mRNA export. The nup85ΔC and nup85ΔN mutants as well as a wild-type control (NUP85 +) were grown for 3 h at 37°C, fixed, and hybridized with a oligo(dT)-FITC probe to reveal poly(A)+ RNA distribution. Cells were also stained with Hoechst 33258 (DNA).
Figure 5
Purification of a native Nup84p complex. (a) TEV protease induced release of affinity-purified ProtA-TEV-Nup85p from IgG-Sepharose beads. Schematic representation of the ProtA-TEV-Nup85p fusion protein consisting of two IgG binding domains (ProtA), a 10 residue long spacer sequence, the 7 residue long TEV cleavage sequence (underlined) with the cleavage site (indicated by an arrow), followed by the Nup85p sequence. After TEV elution from the beads, a first and second eluate (E1 and E2) were obtained. Finally, a low pH eluate (HAc) was collected. All eluates were analyzed by SDS-PAGE and Western blotting using anti-Nup85p and anti-ProtA antibodies. The positions of the ProtA-TEV-Nup85p and the cleaved Nup85p, lacking the ProtA tag are indicated. (b) Gel filtration chromatography of the affinity-purified and TEV-released Nup84p complex. The eluates E1 and E2, which were combined and concentrated to yield the load fraction (L) were analyzed by FPLC Superose 6 column chromatography. 0.6-ml fractions were collected and 25 μl of fractions 1–27, E1 and E2, and the load (L, 5% of the injected material) were analyzed by SDS-PAGE and silverstaining. The six subunits containing Nup84p complex peak in fraction 9, the Nup85p-Seh1p heterodimer in fraction 15. Fractions 9 and 15 were also analyzed by Western blotting using anti-Nup85p, anti-Nup84p, anti-Seh1p, and anti-Sec13p antibodies, respectively. The positions of the components of the Nup84p complex are indicated. Heavy and light IgG chains are evident in fractions 15 and 16, and the TEV protease in fractions 19 and 20. The Superose 6 column was calibrated with several molecular weight marker proteins of 670, 440, 252, and 158 kD (Bio-Rad).
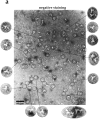
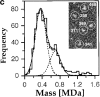
Figure 6
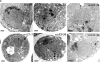
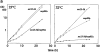
The highly purified Nup84p complex displays a Y-shaped, triskelion-like structure with a molecular mass of ∼375 kD. Electron microscopic appearance of the highly purified Nup84p complex after negative staining (a) and glycerol/rotary metal shadowing (b). In a, a large overview picture is shown, on which Y-shaped structures are highlighted by circles. Selected Y-shaped molecules from this picture were further processed by cut and paste and magnification using Adobe Photoshop, and are shown at the edge of the overview picture. In b, overview pictures (I) and a gallery of selected Nup84p particles (II) are shown. For comparison, shadowed IgG particles are also depicted in b. (c) Mass determination of the highly purified Nup84p complex by STEM. Histogram displaying the mass data determined for 619 particles. Correction has been made for beam-induced mass loss. The two Gaussian curves fall at 368 ± 122 kD (n = 438) and 698 ± 123 kD (n = 158), respectively. The number of particles giving rise to the peaks (n) was estimated by measuring up to and away from the point of peak overlap. The inset reveals a field from an annular dark-field image recorded of an unstained freeze-dried Nup84p sample.
Figure 6
The highly purified Nup84p complex displays a Y-shaped, triskelion-like structure with a molecular mass of ∼375 kD. Electron microscopic appearance of the highly purified Nup84p complex after negative staining (a) and glycerol/rotary metal shadowing (b). In a, a large overview picture is shown, on which Y-shaped structures are highlighted by circles. Selected Y-shaped molecules from this picture were further processed by cut and paste and magnification using Adobe Photoshop, and are shown at the edge of the overview picture. In b, overview pictures (I) and a gallery of selected Nup84p particles (II) are shown. For comparison, shadowed IgG particles are also depicted in b. (c) Mass determination of the highly purified Nup84p complex by STEM. Histogram displaying the mass data determined for 619 particles. Correction has been made for beam-induced mass loss. The two Gaussian curves fall at 368 ± 122 kD (n = 438) and 698 ± 123 kD (n = 158), respectively. The number of particles giving rise to the peaks (n) was estimated by measuring up to and away from the point of peak overlap. The inset reveals a field from an annular dark-field image recorded of an unstained freeze-dried Nup84p sample.
Figure 6
The highly purified Nup84p complex displays a Y-shaped, triskelion-like structure with a molecular mass of ∼375 kD. Electron microscopic appearance of the highly purified Nup84p complex after negative staining (a) and glycerol/rotary metal shadowing (b). In a, a large overview picture is shown, on which Y-shaped structures are highlighted by circles. Selected Y-shaped molecules from this picture were further processed by cut and paste and magnification using Adobe Photoshop, and are shown at the edge of the overview picture. In b, overview pictures (I) and a gallery of selected Nup84p particles (II) are shown. For comparison, shadowed IgG particles are also depicted in b. (c) Mass determination of the highly purified Nup84p complex by STEM. Histogram displaying the mass data determined for 619 particles. Correction has been made for beam-induced mass loss. The two Gaussian curves fall at 368 ± 122 kD (n = 438) and 698 ± 123 kD (n = 158), respectively. The number of particles giving rise to the peaks (n) was estimated by measuring up to and away from the point of peak overlap. The inset reveals a field from an annular dark-field image recorded of an unstained freeze-dried Nup84p sample.
Similar articles
- A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores.
Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. Siniossoglou S, et al. Cell. 1996 Jan 26;84(2):265-75. doi: 10.1016/s0092-8674(00)80981-2. Cell. 1996. PMID: 8565072 - Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins.
Teixeira MT, Siniossoglou S, Podtelejnikov S, Bénichou JC, Mann M, Dujon B, Hurt E, Fabre E. Teixeira MT, et al. EMBO J. 1997 Aug 15;16(16):5086-97. doi: 10.1093/emboj/16.16.5086. EMBO J. 1997. PMID: 9305650 Free PMC article. - Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif.
Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Fabre E, et al. Cell. 1994 Jul 29;78(2):275-89. doi: 10.1016/0092-8674(94)90297-6. Cell. 1994. PMID: 8044840 - The nuclear pore complex.
Davis LI. Davis LI. Annu Rev Biochem. 1995;64:865-96. doi: 10.1146/annurev.bi.64.070195.004245. Annu Rev Biochem. 1995. PMID: 7574503 Review. - Molecular dissection of the nuclear pore complex.
Panté N, Aebi U. Panté N, et al. Crit Rev Biochem Mol Biol. 1996 Apr;31(2):153-99. doi: 10.3109/10409239609106583. Crit Rev Biochem Mol Biol. 1996. PMID: 8740526 Review.
Cited by
- The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth.
Dultz E, Wojtynek M, Medalia O, Onischenko E. Dultz E, et al. Cells. 2022 Apr 25;11(9):1456. doi: 10.3390/cells11091456. Cells. 2022. PMID: 35563762 Free PMC article. Review. - Dissecting the Structural Dynamics of the Nuclear Pore Complex.
Hakhverdyan Z, Molloy KR, Keegan S, Herricks T, Lepore DM, Munson M, Subbotin RI, Fenyö D, Aitchison JD, Fernandez-Martinez J, Chait BT, Rout MP. Hakhverdyan Z, et al. Mol Cell. 2021 Jan 7;81(1):153-165.e7. doi: 10.1016/j.molcel.2020.11.032. Epub 2020 Dec 16. Mol Cell. 2021. PMID: 33333016 Free PMC article. - Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex.
Enninga J, Levay A, Fontoura BM. Enninga J, et al. Mol Cell Biol. 2003 Oct;23(20):7271-84. doi: 10.1128/MCB.23.20.7271-7284.2003. Mol Cell Biol. 2003. PMID: 14517296 Free PMC article. - The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly.
Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. Orjalo AV, et al. Mol Biol Cell. 2006 Sep;17(9):3806-18. doi: 10.1091/mbc.e05-11-1061. Epub 2006 Jun 28. Mol Biol Cell. 2006. PMID: 16807356 Free PMC article. - Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly.
Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Lau CK, et al. Mol Biol Cell. 2009 Sep;20(18):4043-58. doi: 10.1091/mbc.e09-02-0152. Epub 2009 Jul 29. Mol Biol Cell. 2009. PMID: 19641022 Free PMC article.
References
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N.R., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. COPIIa membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases