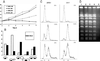Chromosome condensation factor Brn1p is required for chromatid separation in mitosis - PubMed (original) (raw)
Chromosome condensation factor Brn1p is required for chromatid separation in mitosis
I I Ouspenski et al. Mol Biol Cell. 2000 Apr.
Free PMC article
Abstract
This work describes BRN1, the budding yeast homologue of Drosophila Barren and Xenopus condensin subunit XCAP-H. The Drosophila protein is required for proper chromosome segregation in mitosis, and Xenopus protein functions in mitotic chromosome condensation. Mutant brn1 cells show a defect in mitotic chromosome condensation and sister chromatid separation and segregation in anaphase. Chromatid cohesion before anaphase is properly maintained in the mutants. Some brn1 mutant cells apparently arrest in S-phase, pointing to a possible function for Brn1p at this stage of the cell cycle. Brn1p is a nuclear protein with a nonuniform distribution pattern, and its level is up-regulated at mitosis. Temperature-sensitive mutations of BRN1 can be suppressed by overexpression of a novel gene YCG1, which is homologous to another Xenopus condensin subunit, XCAP-G. Overexpression of SMC2, a gene necessary for chromosome condensation, and a homologue of the XCAP-E condensin, does not suppress brn1, pointing to functional specialization of components of the condensin complex.
Figures
Figure 1
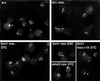
Mitotic chromosome condensation defect in brn1 mutant cells. Examples of rDNA array morphologies, visualized by fluorescence in situ hybridization, are shown. BRN1 (w.t.) or brn1–60 cells were incubated at indicated temperatures for 3.5 h with or without nocodazole (noc). rDNA morphology in smc2–8 cells is shown for comparison. The bottom right panel (noc–>1h 37C) shows brn1–60 cells blocked in nocodazole at permissive temperature for 2.5 h, followed by a shift to 37°C in the continued presence of nocodazole. Arrowheads point to examples of rDNA morphology scored as “intermediate.” The graph below shows percentages of cells with the indicated rDNA morphologies after 3.5 h at the restrictive temperature in the presence of nocodazole. Blind scoring of at least 100 cells in each preparation was performed. Cells that could not be unequivocally assigned to one of the two classes were scored as intermediate (see arrowheads above).
Figure 1
Mitotic chromosome condensation defect in brn1 mutant cells. Examples of rDNA array morphologies, visualized by fluorescence in situ hybridization, are shown. BRN1 (w.t.) or brn1–60 cells were incubated at indicated temperatures for 3.5 h with or without nocodazole (noc). rDNA morphology in smc2–8 cells is shown for comparison. The bottom right panel (noc–>1h 37C) shows brn1–60 cells blocked in nocodazole at permissive temperature for 2.5 h, followed by a shift to 37°C in the continued presence of nocodazole. Arrowheads point to examples of rDNA morphology scored as “intermediate.” The graph below shows percentages of cells with the indicated rDNA morphologies after 3.5 h at the restrictive temperature in the presence of nocodazole. Blind scoring of at least 100 cells in each preparation was performed. Cells that could not be unequivocally assigned to one of the two classes were scored as intermediate (see arrowheads above).
Figure 2
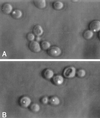
Sister chromatid cohesion is normal in brn1 mutant cells. The centromeric region of chromosome IV in these strains is tagged with an array of Lac operator repeats and visualized by expressing LacI::GFP fusion protein. GFP fluorescence (bright dots) is shown overlaid onto DIC images of cells. Wild-type (A) and _brn1_-60 mutant (B) cells were incubated at 37°C for 3 h in the presence of nocodazole. The two sister centromeres appear as a single dot in >90% of wild-type and mutant cells under these conditions (200 cells of each type scored).
Figure 3
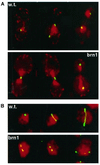
(A) Chromatid separation and segregation in BRN1 (w.t.) and brn1–60 mutant cells. Centromere of chromosome IV, visualized with LacI::GFP, is shown in green; cell nuclei, stained with DAPI (pseudocolored), are bright red; the outline of the cells is visible as red background. Exponentially growing cultures were shifted to 37°C for 3 h and processed for microscopy. (B) Examples of mitotic spindle morphology in BRN1 (w.t.) and brn1–60 cells incubated at 37°C for 3 h. Antitubulin immunofluorescence is shown in green; DNA stained with DAPI is pseudocolored red.
Figure 4
The effect of brn1 mutation on cell cycle progression. (A) Relative viability of _brn1_-60 cells. Midlog phase cultures were split into two halves, and one half was shifted to the restrictive temperature (37°C). At indicated time points, portions of each culture were plated out at a density of 50–500 cells per plate, and the numbers of resulting colonies, divided by the corresponding number of colonies at time zero, were plotted on the graph. (B) Cell cycle stage distribution (% of total; 200 cells scored) of brn1–60 cells at the restrictive temperature, compared with wild-type (BRN1) cells. Asynchronous cultures were incubated at 37°C for 3 h. (C) DNA content of BRN1 and _brn1_-60 cells arrested at the G1 phase with α-factor and released at the restrictive temperature. Samples of cells were fixed at the indicated time points, stained with propidium iodide, and analyzed by flow cytometry. (D) Pulse-field electrophoresis of chromosomal DNA of BRN1 (lanes 1, 2) and brn1–60 mutant cells (lanes 3, 4) grown at 23°C (lanes 1, 3) or incubated at 37°C for 3 h. Lane 5, BRN1 cells treated with 100 mM hydroxyurea for 3 h.
Figure 5
Physical association between Brn1p and Smc2p. Total protein extracts from cells expressing HA-Brn1p (from GAL1 promoter on a CEN plasmid) and Myc-Smc2p (from endogenous promoter on a 2-μm plasmid) were immunoprecipitated with anti-Myc antibody (9E10) and analyzed by immunoblotting with anti-HA and anti-Myc antibodies, as indicated. WCE, Whole cell extract (input), 20 μl; Myc-IP, immunoprecipitation from 400 μl of WCE.
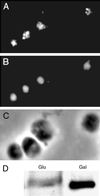
Figure 6
Intracellular localization of the Brn1p protein. (A) Immunofluorescent staining of cells overexpressing Brn1p from the GAL1 promoter on a CEN plasmid, using an affinity-purified anti-Brn1p antibody. (B) Nuclear DNA of the same cells stained with DAPI. (C) Phase-contrast image of the same cells. (D) Levels of Brn1p under induced (Gal) and uninduced (Glu) conditions.
Figure 7
Cell cycle regulation of Brn1p. (A) Levels of endogenous Brn1p in wild-type cells blocked in mitosis with nocodazole (N), or in G1 with α-factor (α), detected by immunoblotting with anti-Brn1p antibody. Equal amounts of total protein were loaded in the two lanes. (B) Brn1p in wild-type cells released from hydroxyurea arrest. Cells were collected every 15 min, and total protein extracts obtained from identical culture volumes were loaded onto each lane. Most cells have completed mitosis (78% unbudded cells, n = 200) 1 h after release, whereas at the 2 h point most cells were in G2 or M (73% cells with bud size more than one-half of the mother cell, n = 200).
Similar articles
- Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren.
Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C. Lavoie BD, et al. Mol Biol Cell. 2000 Apr;11(4):1293-304. doi: 10.1091/mbc.11.4.1293. Mol Biol Cell. 2000. PMID: 10749930 Free PMC article. - Cell cycle-dependent expression and nucleolar localization of hCAP-H.
Cabello OA, Eliseeva E, He WG, Youssoufian H, Plon SE, Brinkley BR, Belmont JW. Cabello OA, et al. Mol Biol Cell. 2001 Nov;12(11):3527-37. doi: 10.1091/mbc.12.11.3527. Mol Biol Cell. 2001. PMID: 11694586 Free PMC article. - A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis.
Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE. Steffensen S, et al. Curr Biol. 2001 Mar 6;11(5):295-307. doi: 10.1016/s0960-9822(01)00096-3. Curr Biol. 2001. PMID: 11267866 - Shaping the metaphase chromosome: coordination of cohesion and condensation.
Losada A, Hirano T. Losada A, et al. Bioessays. 2001 Oct;23(10):924-35. doi: 10.1002/bies.1133. Bioessays. 2001. PMID: 11598959 Review. - Sister chromatid cohesion and recombination in meiosis.
van Heemst D, Heyting C. van Heemst D, et al. Chromosoma. 2000;109(1-2):10-26. doi: 10.1007/s004120050408. Chromosoma. 2000. PMID: 10855491 Review.
Cited by
- Transcriptional homogenization of rDNA repeats in the episome-based nucleolus induces genome-wide changes in the chromosomal distribution of condensin.
Wang BD, Strunnikov A. Wang BD, et al. Plasmid. 2008 Jan;59(1):45-53. doi: 10.1016/j.plasmid.2007.09.003. Epub 2007 Nov 19. Plasmid. 2008. PMID: 18023874 Free PMC article. - Global analysis of cdc14 dephosphorylation sites reveals essential regulatory role in mitosis and cytokinesis.
Kao L, Wang YT, Chen YC, Tseng SF, Jhang JC, Chen YJ, Teng SC. Kao L, et al. Mol Cell Proteomics. 2014 Feb;13(2):594-605. doi: 10.1074/mcp.M113.032680. Epub 2013 Dec 7. Mol Cell Proteomics. 2014. PMID: 24319056 Free PMC article. - NCAPH plays important roles in human colon cancer.
Yin L, Jiang LP, Shen QS, Xiong QX, Zhuo X, Zhang LL, Yu HJ, Guo X, Luo Y, Dong J, Kong QP, Yang CP, Chen YB. Yin L, et al. Cell Death Dis. 2017 Mar 16;8(3):e2680. doi: 10.1038/cddis.2017.88. Cell Death Dis. 2017. PMID: 28300828 Free PMC article. - Condensin is required for chromosome arm cohesion during mitosis.
Lam WW, Peterson EA, Yeung M, Lavoie BD. Lam WW, et al. Genes Dev. 2006 Nov 1;20(21):2973-84. doi: 10.1101/gad.1468806. Genes Dev. 2006. PMID: 17079686 Free PMC article. - C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis.
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. Hagstrom KA, et al. Genes Dev. 2002 Mar 15;16(6):729-42. doi: 10.1101/gad.968302. Genes Dev. 2002. PMID: 11914278 Free PMC article.
References
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
- Cabello OA, Baldini A, Bhat MA, Bellen HJ, Belmont JW. Localization of BRRN-1, the human homologue of D. melanogaster barr to 2q11.2. Genomics. 1997;46:311–313. - PubMed
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Chue C. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–2576. - PubMed
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases