Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31 - PubMed (original) (raw)
Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31
Y E Chang et al. J Virol. 2000 May.
Abstract
Human papillomaviruses (HPVs) infect keratinocytes and induce proliferative lesions. In infected cells, viral gene products alter the activities of cellular proteins, such as Rb and p53, resulting in altered cell cycle response. It is likely that HPV gene products also alter expression of cellular genes. In this study we used microarray analysis to examine the global changes in gene expression induced by high-risk HPV type 31 (HPV31). Among 7,075 known genes and ESTs (expressed sequence tags) tested, we found that 178 were upregulated and 150 were downregulated twofold or more in HPV31 cells compared to normal human keratinocytes. While no specific pattern could be deduced from the list of genes that were upregulated, downregulated genes could be classified to three groups: genes that are involved in the regulation of cell growth, genes that are specifically expressed in keratinocytes, and genes whose expression is increased in response to interferon stimulation. The basal level of expression of several interferon-responsive genes was found to be downregulated in HPV31 cells by both microarray analysis and Northern blot analysis in different HPV31 cell lines. When cells were treated with alpha or gamma interferon, expression of interferon-inducible genes was impaired. At high doses of interferon, the effects were less pronounced. Among the genes repressed by HPV31 was the signal transducer and activator of transcription (Stat-1), which plays a major role in mediating the interferon response. Suppression of Stat-1 expression may contribute to a suppressed response to interferon as well as immune evasion.
Figures
FIG. 1
Global comparison of gene expression in HPV31 and NHKs. Each dot corresponds to the Cy3 (x axis) and Cy5 (y axis) fluorescent intensity of one single element on the microarray. Twofold, 5-fold, and 10-fold changes in expression are indicated as parallel lines. Dots that represent more than 11.5-fold changes are internal controls for hybridization.
FIG. 2
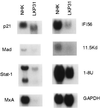
Level of expression in genes suppressed in HPV31 cells demonstrated by microarray analysis is confirmed by Northern blot analysis. Total RNAs (20 μg) from NHKs or LKP31 cells were separated by formaldehyde agarose electrophoresis, transferred to a membrane, and hybridized with various cDNA probes. The slightly faster mobility of p21 and Stat-1 signals in LKP31 cells is likely due to electrophoretic artifact since it was not observed in separate experiments.
FIG. 3
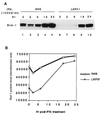
Induction of MxA RNA by IFN-α is impaired in HPV31 cells. (A) Northern blot analysis of 8 μg of total RNA from NHK or LKP31 cells treated with IFN-α (100 or 500 U/ml) with MxA cDNA as a probe. MxA RNA signals in panel A were quantified by a PhosphorImager, and the results are shown graphically in panel B. The induction of MxA RNA by IFN-α reaches comparable levels in LKP31 cells and NHKs at a higher dose (1,000 U/ml) and longer time exposure. (C) Northern blot analysis of 10 μg of total RNA from NHKs and LKP31 cells treated with IFN-α with MxA cDNA as a probe. MxA RNA signals in panel C were quantified by a PhosphorImager, and the results are shown graphically in panel D.
FIG. 4
The induction of Stat-1 protein by IFN-α is impaired in HPV31 cells. (A) Western blot analysis of equal amounts of protein lysates from NHKs and LKP31 cells treated with IFN-α (100 or 500 U/ml) to detect Stat-1 protein level. Stat-1 protein migrates in SDS-PAGE as a doublet with molecular weights of 91,000 and 84,000. Quantitative estimation of the results in panel A was done with a densitometer and shown graphically in panel B. The induction of Stat-1 protein by IFN-α reaches comparable levels in LKP31 cells and NHKs with a higher dose (1,000 U/ml) and longer time. (C) Western blot analysis of equal amount of protein lysates from NHKs or LKP31 cells treated with IFN-α to detect Stat-1 protein. Quantitative estimation of the results in panel C was done with a densitometer and shown graphically in panel D.
FIG. 5
The induction of Stat-1 protein by IFN-γ is impaired in HPV31 cells. (A) Western blot analysis of equal amounts of protein lysates from NHKs and LKP31 cells treated with IFN-γ (1,000 U/ml) to detect Stat-1 protein level. The image in panel A was quantified by a densitometer, and the results are shown graphically in panel B.
Similar articles
- Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes.
Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Nees M, et al. J Virol. 2001 May;75(9):4283-96. doi: 10.1128/JVI.75.9.4283-4296.2001. J Virol. 2001. PMID: 11287578 Free PMC article. - The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes.
Stubenrauch F, Hummel M, Iftner T, Laimins LA. Stubenrauch F, et al. J Virol. 2000 Feb;74(3):1178-86. doi: 10.1128/jvi.74.3.1178-1186.2000. J Virol. 2000. PMID: 10627528 Free PMC article. - Global changes in STAT target selection and transcription regulation upon interferon treatments.
Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M. Hartman SE, et al. Genes Dev. 2005 Dec 15;19(24):2953-68. doi: 10.1101/gad.1371305. Epub 2005 Nov 30. Genes Dev. 2005. PMID: 16319195 Free PMC article. - An indoleamine 2,3-dioxygenase-negative mutant is defective in stat1 DNA binding: differential response to IFN-gamma and IFN-alpha.
Sotero-Esteva WD, Wolfe D, Ferris M, Taylor MW. Sotero-Esteva WD, et al. J Interferon Cytokine Res. 2000 Jul;20(7):623-32. doi: 10.1089/107999000414790. J Interferon Cytokine Res. 2000. PMID: 10926204
Cited by
- Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes.
Sandberg A, Lindell G, Källström BN, Branca RM, Danielsson KG, Dahlberg M, Larson B, Forshed J, Lehtiö J. Sandberg A, et al. Mol Cell Proteomics. 2012 Jul;11(7):M112.016998. doi: 10.1074/mcp.M112.016998. Epub 2012 Apr 12. Mol Cell Proteomics. 2012. PMID: 22499770 Free PMC article. - Progress in the application of DNA microarrays.
Lobenhofer EK, Bushel PR, Afshari CA, Hamadeh HK. Lobenhofer EK, et al. Environ Health Perspect. 2001 Sep;109(9):881-91. doi: 10.1289/ehp.01109881. Environ Health Perspect. 2001. PMID: 11673116 Free PMC article. Review. - The small non-coding RNA response to virus infection in the Leishmania vector Lutzomyia longipalpis.
Ferreira FV, Aguiar ERGR, Olmo RP, de Oliveira KPV, Silva EG, Sant'Anna MRV, Gontijo NF, Kroon EG, Imler JL, Marques JT. Ferreira FV, et al. PLoS Negl Trop Dis. 2018 Jun 4;12(6):e0006569. doi: 10.1371/journal.pntd.0006569. eCollection 2018 Jun. PLoS Negl Trop Dis. 2018. PMID: 29864168 Free PMC article. - Inactivation of p53 rescues the maintenance of high risk HPV DNA genomes deficient in expression of E6.
Lorenz LD, Rivera Cardona J, Lambert PF. Lorenz LD, et al. PLoS Pathog. 2013 Oct;9(10):e1003717. doi: 10.1371/journal.ppat.1003717. Epub 2013 Oct 24. PLoS Pathog. 2013. PMID: 24204267 Free PMC article. - IPA Analysis of Cervicovaginal Fluid from Precancerous Women Points to the Presence of Biomarkers for the Precancerous State of Cervical Carcinoma.
Van Ostade X, Dom M, Van Raemdonck G. Van Ostade X, et al. Proteomes. 2014 Aug 13;2(3):426-450. doi: 10.3390/proteomes2030426. Proteomes. 2014. PMID: 28250389 Free PMC article.
References
- Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. - PubMed
- Barnard P, McMillan N A J. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-α. Virology. 1999;259:305–313. - PubMed
- Cheng S, Schmidt G D, Murant T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous