MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery - PubMed (original) (raw)
MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery
N K Kaludov et al. Nucleic Acids Res. 2000.
Abstract
The pathways for selective transcriptional repression of methylated DNA templates by the methyl-CpG-binding protein MeCP2 have been investigated using a purified in vitro transcription system that does not assemble chromatin. MeCP2 selectively inhibits transcription complex assembly on methylated DNA but does not destabilize a pre-assembled transcription complex. MeCP2 functions to repress transcription at a distance of >500 bp from the transcription start site. The transcription repression domain (TRD) of MeCP2 will repress transcription in vitro when fused to a heterologous Gal4 DNA-binding domain. The TRD associates with TFIIB. Exogenous TFIIB does not relieve transcriptional repression established by either intact MeCP2 or a Gal4-TRD fusion protein under these in vitro conditions, nor does the addition of histone deacetylase inhibitors. We find that the transcriptional repression established by both MeCP2 and the Gal4-TRD fusion protein in vitro also correlates with selective assembly of large nucleoprotein complexes. The formation of such complexes reflects a local concentration of DNA-bound transcriptional repressor that may stabilize a state of repression even in the presence of exogenous transcriptional machinery.
Figures
Figure 1
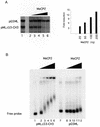
MeCP2-responsive in vitro transcription assay. (A) Transcription reactions were as described (28) except that one of the G-less cassette templates pMLΔ53-CH3 was _Sss_I methylated, whereas pG5ML remained unmethylated. Recombinant MeCP2 was preincubated with the templates for 20 min at room temperature prior to addition of the basal transcription factors (IIB, IIE, IIF, TBP, IIH and polymerase II). The reaction was allowed to continue for 60 min at 30°C. The RNAs were extracted and run on a denaturing gel. (Left) Lane 1, standard transcription reaction, no MeCP2; lanes 2–6, standard transcription reaction preincubated with 10, 20, 50, 100 and 200 ng of MeCP2, respectively. (Right) Quantitation of transcription from the pMLΔ53-CH3 template relative to pG5ML expressed as fold reduction relative to MeCP2 mass per reaction. (B) MeCP2 was preincubated with the DNA templates as in the transcription assay. The complexes were resolved on a 0.7% agarose gel. Lanes 1–6, methylated and labeled pMLΔ53 template; lanes 7–12, labeled pG5ML template; lanes 1 and 7, no MeCP2; lanes 2 and 8, 10 ng; lanes 3 and 9, 20 ng; lanes 4 and 10, 50 ng; lanes 5 and 11, 100 ng; lanes 6 and 12, 200 ng.
Figure 2
MeCP2 can repress transcription from a distance. (A) Six _Hha_I sites were cloned next to the adenovirus major late promoter, 250 and 500 bp away from the start site of transcription +1. The templates were methylated with _Hha_I which resulted in a methylation cluster (black box). Other methylated CpG islands are indicated by vertical bars. The templates were assayed in a standard transcription reaction. (B) Lanes 1–3, methylated _Sss_I template; lanes 4–6, methylated _Hha_I template with six _Hha_I sites next to the promoter; lanes 7–9, six _Hha_I sites 250 bp away; lanes 10–12, six _Hha_I sites 500 bp away; lanes 1, 4, 7 and 10, no MeCP2; lanes 2, 5, 8 and 11, 100 ng; lanes 3, 6, 9 and 12, 200 ng. (C) Quantitation of data shown in (A) as in Figure 1A. (D) Agarose gel shift with the same DNA templates as in (A). Lanes 1, 5, 9 and 13, no addition of MeCP2; lanes 2, 6, 10 and 14, 20 ng; lanes 3, 7, 11 and 15; 100 ng; lanes 4, 8, 12 and 16, 200 ng. (E) The methylated adenovirus major late promoter is accessible for restriction nuclease digestion in the presence of MeCP2. Methylated _Sss_I or _Hha_I template as indicated [labeled as in (A)] was incubated exactly as in the standard transcription reaction in the presence (+) or absence (–) of 200 ng MeCP2 for 15 min at room temperature as indicated. _Ear_I (2 U/reaction) was added and the incubation was continued for 45 min at 30°C. Reactions were deproteinated with phenol/chloroform, the DNA was ethanol precipitated and then the fragments were separated on a 1.2% agarose gel and the extent of digestion was analyzed by Southern blotting. The probe was ML64, a 64 bp oligonucleotide that spans the promoter region of the adenovirus major late promoter, end-labeled with T4 polynucleotide kinase. The positions of uncut and cut (restricted) DNA are indicated.
Figure 2
MeCP2 can repress transcription from a distance. (A) Six _Hha_I sites were cloned next to the adenovirus major late promoter, 250 and 500 bp away from the start site of transcription +1. The templates were methylated with _Hha_I which resulted in a methylation cluster (black box). Other methylated CpG islands are indicated by vertical bars. The templates were assayed in a standard transcription reaction. (B) Lanes 1–3, methylated _Sss_I template; lanes 4–6, methylated _Hha_I template with six _Hha_I sites next to the promoter; lanes 7–9, six _Hha_I sites 250 bp away; lanes 10–12, six _Hha_I sites 500 bp away; lanes 1, 4, 7 and 10, no MeCP2; lanes 2, 5, 8 and 11, 100 ng; lanes 3, 6, 9 and 12, 200 ng. (C) Quantitation of data shown in (A) as in Figure 1A. (D) Agarose gel shift with the same DNA templates as in (A). Lanes 1, 5, 9 and 13, no addition of MeCP2; lanes 2, 6, 10 and 14, 20 ng; lanes 3, 7, 11 and 15; 100 ng; lanes 4, 8, 12 and 16, 200 ng. (E) The methylated adenovirus major late promoter is accessible for restriction nuclease digestion in the presence of MeCP2. Methylated _Sss_I or _Hha_I template as indicated [labeled as in (A)] was incubated exactly as in the standard transcription reaction in the presence (+) or absence (–) of 200 ng MeCP2 for 15 min at room temperature as indicated. _Ear_I (2 U/reaction) was added and the incubation was continued for 45 min at 30°C. Reactions were deproteinated with phenol/chloroform, the DNA was ethanol precipitated and then the fragments were separated on a 1.2% agarose gel and the extent of digestion was analyzed by Southern blotting. The probe was ML64, a 64 bp oligonucleotide that spans the promoter region of the adenovirus major late promoter, end-labeled with T4 polynucleotide kinase. The positions of uncut and cut (restricted) DNA are indicated.
Figure 3
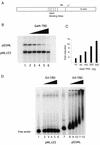
MeCP2 interferes with preinitiation complex (PIC) formation/function. (A) Lane 1, standard transcription reaction, no MeCP2; lane 2, 200 ng MeCP2 preincubated with the templates for 20 min prior to adding the transcription factors (–20 min); lane 3, 200 ng MeCP2 added at the same time as the transcription factors (0 min); lanes 4–6, 200 ng MeCP2 added 20, 40 and 55 min after addition of the transcription factors, respectively. (B) Quantitation of transcription from the pMLΔ53-CH3 template relative to pG5ML expressed as fold reduction relative to MeCP2 mass per reaction.
Figure 4
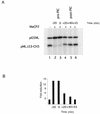
Contacts of MeCP2 with the basal transcription machinery. (A) MeCP2 interacts with TFIIB in a GST pull-down assay. GST–MeCP2 was incubated with recombinant IIA, IIB, IIEα, IIF and purified HeLa IID, IIH and RNA polymerase II in the same buffer conditions as in the transcription reaction (Materials and Methods). GST–agarose was added to the reactions and incubation was continued for another 2–4 h at 4°C. The beads were washed three times with a buffer containing 500 mM KCl, 0.1% NP-40 and the proteins retained by the agarose were separated by SDS–PAGE, transferred to nitrocellulose and probed with antibodies against the proteins of interest. The three lanes show 20% (one fifth) of input, the eluate from GST alone and the eluate from GST–MeCP2. (B) The TRD of MeCP2 but not the MBD interacts with TFIIB in a GST pull-down assay. As in (A) except that GST–TRD and GST–MBD (Materials and Methods) were used.
Figure 5
Gal4–TRD will repress transcription in cis. (A) The template used in these experiments was pG5ML in which five GAL4-binding sites are positioned 68 bp upstream from the start site of transcription (+1) of the adenovirus major late promoter (ML). (B) The TRD of MeCP2 when thethered to a Gal4-binding domain can repress transcription from a template containing Gal4-binding sites. Reaction conditions were as in Figure 1. Lane 1, standard transcription reaction; lanes 2–6, 50, 100, 200, 500 and 1000 ng of Gal4–TRD was preincubated with the templates for 30 min at room temperature, prior to adding the transcription factors. (C) Quantitation of transcription from the pG5ML template relative to pMLΔ53 template expressed as fold reduction relative to Gal4–TRD mass per reaction. (D) Gal4–TRD protein aggregates its target template. Gal4–TRD was preincubated with the DNA templates as in the transcription assay. The complexes were resolved in a 0.7% agarose gel. Lanes 1–6, labeled pMLΔ53 template; lanes 7–12, labeled pG5ML template; lanes 1 and 7, no addition of Gal4–TRD; lanes 2 and 8; 50 ng; lanes 3 and 9, 100 ng; lanes 4 and 10, 200 ng; lanes 5 and 11, 500 ng; lanes 6 and 12, 1000 ng.
Figure 6
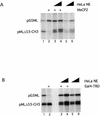
HeLa nuclear extract can rescue MeCP2- and Gal4–TRD-induced repression. (A) Increasing amounts of HeLa nuclear extract (1 and 5 µg, respectively) were added to the standard transcription reaction in the presence (lanes 3 and 4) or absence (lanes 5 and 6) of 200 ng MeCP2. As controls lane 1 contains no MeCP2 and no nuclear extract, lane 2 contains 200 ng MeCP2 and no nuclear extract. (B) As (A) except that Gal4–TRD (1 µg) was used as repressor.
Similar articles
- Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex.
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nan X, et al. Nature. 1998 May 28;393(6683):386-9. doi: 10.1038/30764. Nature. 1998. PMID: 9620804 - Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription.
Yu F, Zingler N, Schumann G, Strätling WH. Yu F, et al. Nucleic Acids Res. 2001 Nov 1;29(21):4493-501. doi: 10.1093/nar/29.21.4493. Nucleic Acids Res. 2001. PMID: 11691937 Free PMC article. - The Ski protein family is required for MeCP2-mediated transcriptional repression.
Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. Kokura K, et al. J Biol Chem. 2001 Sep 7;276(36):34115-21. doi: 10.1074/jbc.M105747200. Epub 2001 Jul 5. J Biol Chem. 2001. PMID: 11441023 - Methyl-CpG-binding proteins. Targeting specific gene repression.
Ballestar E, Wolffe AP. Ballestar E, et al. Eur J Biochem. 2001 Jan;268(1):1-6. doi: 10.1046/j.1432-1327.2001.01869.x. Eur J Biochem. 2001. PMID: 11121095 Review. - The biological functions of the methyl-CpG-binding protein MeCP2 and its implication in Rett syndrome.
Nan X, Bird A. Nan X, et al. Brain Dev. 2001 Dec;23 Suppl 1:S32-7. doi: 10.1016/s0387-7604(01)00333-3. Brain Dev. 2001. PMID: 11738839 Review.
Cited by
- MiR-422a promotes adipogenesis via MeCP2 downregulation in human bone marrow mesenchymal stem cells.
Giuliani A, Sabbatinelli J, Amatori S, Graciotti L, Silvestrini A, Matacchione G, Ramini D, Mensà E, Prattichizzo F, Babini L, Mattiucci D, Busilacchi EM, Bacalini MG, Espinosa E, Lattanzio F, Procopio AD, Olivieri F, Poloni A, Fanelli M, Rippo MR. Giuliani A, et al. Cell Mol Life Sci. 2023 Feb 27;80(3):75. doi: 10.1007/s00018-023-04719-6. Cell Mol Life Sci. 2023. PMID: 36847916 Free PMC article. - Genomic and regulatory characteristics of significant transcription factors in colorectal cancer metastasis.
Zhou B, Guo R. Zhou B, et al. Sci Rep. 2018 Dec 13;8(1):17836. doi: 10.1038/s41598-018-36168-8. Sci Rep. 2018. PMID: 30546056 Free PMC article. - Rett syndrome and MeCP2: linking epigenetics and neuronal function.
Shahbazian MD, Zoghbi HY. Shahbazian MD, et al. Am J Hum Genet. 2002 Dec;71(6):1259-72. doi: 10.1086/345360. Epub 2002 Nov 19. Am J Hum Genet. 2002. PMID: 12442230 Free PMC article. Review. No abstract available. - Cancer epigenetics: linking basic biology to clinical medicine.
Tsai HC, Baylin SB. Tsai HC, et al. Cell Res. 2011 Mar;21(3):502-17. doi: 10.1038/cr.2011.24. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321605 Free PMC article. Review. - Temporal and epigenetic regulation of neurodevelopmental plasticity.
Allen ND. Allen ND. Philos Trans R Soc Lond B Biol Sci. 2008 Jan 12;363(1489):23-38. doi: 10.1098/rstb.2006.2010. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 17311782 Free PMC article. Review.
References
- Amir R.E., Van den Veyver,I.B., Wan,M., Tran,C.Q., Francke,U. and Zoghbi,H.Y. (1999) Nature Genet., 23, 185–188. - PubMed
- Tate P., Skarnes,W. and Bird,A. (1996) Nature Genet., 12, 205–208. - PubMed
- Baylin S.B. (1999) Semin. Cancer Biol., 9, 327–328. - PubMed
- Bird A.P. and Wolffe,A.P. (1999) Cell, 99, 451–454. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources