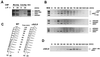Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase - PubMed (original) (raw)
Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase
L Zou et al. Mol Cell Biol. 2000 May.
Abstract
In Saccharomyces cerevisiae, replication origins are activated with characteristic timing during S phase. S-phase cyclin-dependent kinases (S-CDKs) and Cdc7p-Dbf4p kinase are required for origin activation throughout S phase. The activation of S-CDKs leads to association of Cdc45p with chromatin, raising the possibility that Cdc45p defines the assembly of a new complex at each origin. Here we show that both Cdc45p and replication protein A (RPA) bind to Mcm2p at the G(1)-S transition in an S-CDK-dependent manner. During S phase, Cdc45p associates with different replication origins at specific times. The origin associations of Cdc45p and RPA are mutually dependent, and both S-CDKs and Cdc7p-Dbf4p are required for efficient binding of Cdc45p to origins. These findings suggest that S-CDKs and Cdc7p-Dbf4p promote loading of Cdc45p and RPA onto a preformed prereplication complex at each origin with preprogrammed timing. The ARS1 association of Mcm2p, but not that of the origin recognition complex, is diminished by disruption of the B2 element of ARS1, a potential origin DNA-unwinding element. Cdc45p is required for recruiting DNA polymerase alpha onto chromatin, and it associates with Mcm2p, RPA, and DNA polymerase epsilon only during S phase. These results suggest that the complex containing Cdc45p, RPA, and MCMs is involved in origin unwinding and assembly of replication forks at each origin.
Figures
FIG. 1
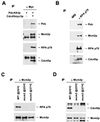
Cdc45p associates with Mcm2p, Polɛ, and RPA p70 in vivo. (A) Mcm2p, PolɛHA3p, and RPA p70 coprecipitate specifically with Cdc45myc3p. Cells expressing (+) both PolɛHA3p and Cdc45myc3p (YB0550) or only PolɛHA3p (OAY618) were arrested in S phase with 0.1 M HU for 2 h. Whole-cell extracts were prepared and subjected to immunoprecipitations (IP) with non-cross-linked anti-myc antibody. ∗, dimers of immunoglobulin G. (B) Mcm2p, PolɛHA3p and Cdc45myc3p coprecipitate specifically with RPA p70. Whole-cell extract was prepared from S-phase wild-type (YB0550) cells. Immunoprecipitations were performed with either normal rabbit serum (NRS) or anti-RPA p70 antibody. (C) RPA p70 coprecipitates with Mcm2p only in wild-type but not in mcm2-1 cell extracts. Wild-type (YB0550) (WT) and mcm2-1 (YB0551) cells were arrested with HU for 2 h at the indicated temperatures. Immunoprecipitations were carried out with anti-Mcm2p antibody cross-linked to protein G. (D) Cdc45HA3p coprecipitates with Mcm2p only in wild-type but not in mcm2-1 cell extracts. Wild-type (YB0469) and mcm2-1 (YB0476) cells expressing Cdc45HA3p were arrested as in panel C, and immunoprecipitations were performed with non-cross-linked anti-Mcm2p antibody. Shown are immunoblots of the proteins present in the immunoprecipitation described above. The immunoblotting antibodies were against the proteins indicated at the right of each panel.
FIG. 2
Cdc45p interacts with Mcm2p, Polɛ, RPA p70, and p34 in a cell cycle-dependent manner. (A) Immunoprecipitation (IP) of Cdc45myc3p across the cell cycle. Wild-type cells expressing both PolɛHA3p and Cdc45myc3p (YB0550) were synchronized in G1 with α-factor and then released into yeast-peptone-dextrose medium at 25°C. The cells were collected at the indicated time points. Whole-cell extracts were prepared and subjected to immunoprecipitations with anti-myc antibody. (B) DNA content of the time point samples used in panel A.
FIG. 3
S-CDK activity is required for the interactions among Cdc45p, Mcm2p, and RPA. (A) Overexpression of Sic1ΔNTp prevents binding of Cdc45p to Mcm2p. Cells bearing GAL1-SIC1ΔNT (YB0553) were arrested with α-factor in raffinose (raf)-containing medium and then released into medium containing either raffinose or galactose (gal) at 25°C. As a control, wild-type (WT) cells (YB0469) that do not carry GAL1-SIC1ΔNT were also synchronized with α-factor in raffinose and released into galactose. Whole-cell extracts were prepared at the indicated time points and were immunoprecipitated with anti-Mcm2p antibody. Cdc45HA3p coprecipitated with Mcm2p was analyzed by immunoblotting. (B) Overexpression of Sic1ΔNTp prevents binding of RPA p70 and p34 to Mcm2p. RPA p70 and p34 coprecipitated with Mcm2p in panel A were analyzed by immunoblotting. (C) DNA content of the samples used in panels A and B.
FIG. 4
Cell cycle regulation of association of Cdc45p with ARS-containing fragments. (A) Cdc45p and Mcm2p associate with origins differently in G1 and early S phase. Wild-type (WT) cells expressing Cdc45HA3p (YB0469) were arrested in G1 with α-factor and then released into yeast-peptone-dextrose medium at 25°C. The cells were collected at the α-factor block or 30 min after release. CHIP analyses of Mcm2p and Cdc45p were performed in parallel. WCE, input DNA prepared from the whole-cell extract; _ARS1_-4kb, a region 4 kb away from ARS1 towards the left telomere. (B) Association of Cdc45p with different origins during the cell cycle. Wild-type cells were synchronized in G1 with α-factor and then released into YPD at 25°C. CHIP analysis of Cdc45p was performed at each time point. +4 kb and −8 kb, regions on both sides of ARS1 that are 4 or 8 kb away. (C) DNA content of the time point samples used in panels B and D. (D) Association of Cdc45p with ARS1 is delayed in the absence of CLB5 and CLB6. YB0477 (clb5,6Δ and CDC45HA3) cells were released from an α-factor block at 25°C. CHIP analysis of Cdc45p was performed as described above.
FIG. 5
Associations of Cdc45p with chromatin, Mcm2p, and ARS1 are affected in cdc7 and dbf4 mutants. (A) Association of Cdc45p with chromatin is reduced in cdc7 and dbf4 mutants. Wild-type (YB0469) (WT), cdc7-1 (YB0472), cdc7-4 (YB0547), and dbf4-1 (YB0548) cells expressing Cdc45HA3p were synchronized with α-factor and released at either 25 or 35°C. The cells were collected at the α-factor block or 30 min after release. Lysates were prepared and subjected to chromatin fractionation. Immunoblots of Cdc45p and Orc3p in the chromatin sediments (P) and Cdc45p in the supernatants (S) are shown. (B) Cdc45p-Mcm2p interaction is reduced in cdc7 and dbf4 mutants at the nonpermissive temperature. Cells were arrested as in panel A and released at 37°C. Whole-cell extracts (WCE) were prepared from the cells collected at the α-factor block or 30 min after release and subjected to immunoprecipitations (IP) using an anti-Mcm2p antibody. (C) Association of Cdc45p with ARS1 is undetectable in cdc7-1, cdc7-4, and dbf4-1 cells. Wild-type and mutant cells were synchronized with α-factor as in panel A and released at 35°C. The cells were collected at the indicated time points and analyzed by Cdc45p CHIP. (D) DNA content of the samples used in panel C.
FIG. 6
Origin associations of Cdc45p and RPA are mutually dependent. (A) ARS1 association of RPA p70 requires Cdc45p. Wild-type (WT) and cdc45-1 (YB0298) cells were synchronized in G1 with α-factor and released into medium containing 0.1 M HU at 13°C. CHIP analysis of RPA p70 was performed at the indicated time points. (B) DNA content of the cells blocked and released as in panel A but in the absence of HU. (C) ARS1 association of Cdc45p requires RPA p34. Wild-type and rfa2-2 (YB0549) cells were synchronized in G1 with α-factor and released at 35°C in the absence of HU. CHIP analysis of Cdc45p was performed at the indicated time points. (D) DNA content of the samples used in panel C. WCE, input DNA from whole-cell extract.
FIG. 7
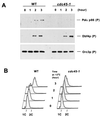
Cdc45p is required for loading Polα p86 but not Dbf4p onto chromatin. (A) Wild-type (K6388) (WT) and cdc45-1 (YB0552) cells were synchronized in G1 with α-factor and released at 13°C. Cells were collected at the indicated time points and were processed for chromatin fractionation. The chromatin-sediment fractions (P) were analyzed by immunoblotting. (B) DNA content of the samples used in panel A.
FIG. 8
The B2 element of ARS1 is involved in association with Mcm2p but not ORC. The Cdc45HA3p-expressing cells with the chromosomal ARS1 mutated in either the A (YB0576), B1 (YB0577), B2 (YB0578), or B3 (YB0579) element were analyzed by CHIP assays. Asynchronously growing wild-type (WT) and ARS1 mutant cells were subjected to CHIP analysis with antibodies against Orc2p-Orc3p (antibodies against Orc2p and Orc3p were mixed and used together) and Mcm2p, respectively.
Similar articles
- Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin.
Zou L, Stillman B. Zou L, et al. Science. 1998 Apr 24;280(5363):593-6. doi: 10.1126/science.280.5363.593. Science. 1998. PMID: 9554851 - Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation.
Nougarède R, Della Seta F, Zarzov P, Schwob E. Nougarède R, et al. Mol Cell Biol. 2000 Jun;20(11):3795-806. doi: 10.1128/MCB.20.11.3795-3806.2000. Mol Cell Biol. 2000. PMID: 10805723 Free PMC article. - Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases.
Tanaka T, Nasmyth K. Tanaka T, et al. EMBO J. 1998 Sep 1;17(17):5182-91. doi: 10.1093/emboj/17.17.5182. EMBO J. 1998. PMID: 9724654 Free PMC article. - First the CDKs, now the DDKs.
Johnston LH, Masai H, Sugino A. Johnston LH, et al. Trends Cell Biol. 1999 Jul;9(7):249-52. doi: 10.1016/s0962-8924(99)01586-x. Trends Cell Biol. 1999. PMID: 10370238 Review. - Initiation of DNA replication in eukaryotes is an intriguing cascade of protein interactions.
Sharova NP, Abramova EB. Sharova NP, et al. Biochemistry (Mosc). 2002 Nov;67(11):1217-23. doi: 10.1023/a:1021389018998. Biochemistry (Mosc). 2002. PMID: 12495416 Review.
Cited by
- Chromatin-dependent and -independent regulation of DNA replication origin activation in budding yeast.
Lõoke M, Kristjuhan K, Värv S, Kristjuhan A. Lõoke M, et al. EMBO Rep. 2013 Feb;14(2):191-8. doi: 10.1038/embor.2012.196. Epub 2012 Dec 7. EMBO Rep. 2013. PMID: 23222539 Free PMC article. - Local and global functions of Timeless and Tipin in replication fork protection.
Leman AR, Noguchi E. Leman AR, et al. Cell Cycle. 2012 Nov 1;11(21):3945-55. doi: 10.4161/cc.21989. Epub 2012 Sep 17. Cell Cycle. 2012. PMID: 22987152 Free PMC article. Review. - The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release.
Lesur I, Le Provost G, Bento P, Da Silva C, Leplé JC, Murat F, Ueno S, Bartholomé J, Lalanne C, Ehrenmann F, Noirot C, Burban C, Léger V, Amselem J, Belser C, Quesneville H, Stierschneider M, Fluch S, Feldhahn L, Tarkka M, Herrmann S, Buscot F, Klopp C, Kremer A, Salse J, Aury JM, Plomion C. Lesur I, et al. BMC Genomics. 2015 Feb 21;16(1):112. doi: 10.1186/s12864-015-1331-9. BMC Genomics. 2015. PMID: 25765701 Free PMC article. - The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication.
Leman AR, Noguchi E. Leman AR, et al. Genes (Basel). 2013 Mar 1;4(1):1-32. doi: 10.3390/genes4010001. Genes (Basel). 2013. PMID: 23599899 Free PMC article. - Schizosaccharomyces pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding.
Dolan WP, Sherman DA, Forsburg SL. Dolan WP, et al. Chromosoma. 2004 Sep;113(3):145-56. doi: 10.1007/s00412-004-0302-8. Epub 2004 Aug 3. Chromosoma. 2004. PMID: 15338237
References
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. - PubMed
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases