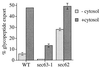The protein translocation channel mediates glycopeptide export across the endoplasmic reticulum membrane - PubMed (original) (raw)
The protein translocation channel mediates glycopeptide export across the endoplasmic reticulum membrane
P Gillece et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Peptides and misfolded secretory proteins are transported efficiently from the endoplasmic reticulum (ER) lumen to the cytosol, where the proteins are degraded by proteasomes. Protein export depends on Sec61p, the ribosome-binding core component of the protein translocation channel in the ER membrane. We found that prebinding of ribosomes abolished export of a glycopeptide from yeast microsomes. Deletion of SSH1, which encodes a ribosome-binding Sec61p homologue in the ER, had no effect on glycopeptide export. A collection of cold-sensitive sec61 mutants displayed a variety of phenotypes: two mutants strongly defective in misfolded protein export from the ER, sec61-32 and sec61-41, displayed only minor peptide export defects. Glycopeptide export was severely impaired, however, in several sec61 mutants that were only marginally defective in misfolded protein export. In addition, a mutation in SEC63 strongly reduced peptide export from the ER. ER-luminal ATP was required for both misfolded protein and glycopeptide export. We conclude that the protein translocation channel in the ER membrane mediates glycopeptide transport across the ER membrane.
Figures
Figure 1
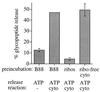
Ribosomes inhibit glycopeptide export from the ER. Yeast cytosol (18 mg/ml) was separated by centrifugation into ribosomes and a ribosome-free supernatant fraction and the ribosomal pellet resuspended in the original volume B88. Wild-type microsomes [RSY255, 2 μl microsomes (_A_280 = 30) per sample] were translocated with 125I-Ac-NYT-NH2, washed, and incubated for 10 min at 24°C in 12.5 μl B88, 12.5 μl B88 containing nontranslating ribosomes, or 12.5 μl ribosome-free cytosol, as indicated. Glycopeptide export was assayed subsequently for 30 min at 24°C in the absence (−cytosol) or presence (all others) of cytosol and ATP, as described in Methods. Samples contained 2–5 × 104 Con A precipitable cpm, were done in duplicate, and the experiment was repeated three times.
Figure 2
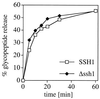
Ssh1p is not required for glycopeptide export. Wild-type (SSH1) and Δ_ssh1_ microsomes were assayed for glycopeptide export as described in Methods. At individual time points, duplicate samples were transferred to ice, the membranes sedimented by centrifugation, and glycopeptide in the supernatatant fraction analyzed by Con A precipitation and γ-counting.
Figure 3
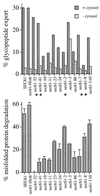
Specific sec61 mutants are defective in glycopeptide export from the ER. Microsomes were prepared from SEC61 wild-type and mutant strains grown at their respective permissive temperatures (see Methods). (Upper) Glycopeptide export. Each strain was assayed in duplicate for glycopeptide export for 30 min at 24°C in the presence or individual absence of ATP and cytosol, as described in Methods. Release at 30 min is shown. Nonspecific release in the absence of ATP was less than 5% and subtracted. Mutants with significantly different effects on glycopeptide and misfolded protein export are marked with asterisks. (Lower) Misfolded protein export. Mutant α-factor precursor (pΔgpαf) was translocated into wild type or sec61 mutant microsomes and Δgpαf export and degradation initiated by the addition of ATP and cytosol, as described in Methods. After 30 min at 24°C, proteins were precipitated with TCA, resolved by gel electrophoresis, and Δgpαf quantified by using a Bio-Rad phosphorimager. Samples were done in duplicate and the experiment repeated twice.
Figure 4
Glycopeptide export from the ER depends on Sec63p. Microsomes were prepared from wild-type and sec62 or sec63 mutant cells grown at the permissive temperature (see Methods) and glycopeptide export assayed as described in Fig. 1.
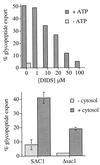
Figure 5
Glycopeptide export across the ER membrane requires ATP in the ER lumen. (Upper) DIDS inhibits glycopeptide export from the ER. Wild-type (RSY255) microsomes were translocated with 125I-Ac-NYT-NH2, washed with B88 containing the indicated concentration of DIDS, and glycopeptide export in the presence of the indicated concentrations of DIDS assayed in duplicate as described in Fig. 1. (Lower) Δ_sac1_ microsomes are defective in glycopeptide export from the ER. Microsomes were prepared from wild-type (SAC1) and Δ_sac1_ cells and glycopeptide export assayed as described in Fig. 1.
Similar articles
- Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation.
Pilon M, Schekman R, Römisch K. Pilon M, et al. EMBO J. 1997 Aug 1;16(15):4540-8. doi: 10.1093/emboj/16.15.4540. EMBO J. 1997. PMID: 9303298 Free PMC article. - The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane.
Kalies KU, Allan S, Sergeyenko T, Kröger H, Römisch K. Kalies KU, et al. EMBO J. 2005 Jul 6;24(13):2284-93. doi: 10.1038/sj.emboj.7600731. Epub 2005 Jun 23. EMBO J. 2005. PMID: 15973433 Free PMC article. - Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane.
Römisch K. Römisch K. J Cell Sci. 1999 Dec;112 ( Pt 23):4185-91. doi: 10.1242/jcs.112.23.4185. J Cell Sci. 1999. PMID: 10564637 Review. - Endoplasmic reticulum-associated degradation.
Römisch K. Römisch K. Annu Rev Cell Dev Biol. 2005;21:435-56. doi: 10.1146/annurev.cellbio.21.012704.133250. Annu Rev Cell Dev Biol. 2005. PMID: 16212502 Review.
Cited by
- N-glycosylation does not affect the catalytic activity of ricin a chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum.
Yan Q, Li XP, Tumer NE. Yan Q, et al. Traffic. 2012 Nov;13(11):1508-21. doi: 10.1111/j.1600-0854.2012.01404.x. Epub 2012 Sep 7. Traffic. 2012. PMID: 22882900 Free PMC article. - Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system.
Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Wahlman J, et al. Cell. 2007 Jun 1;129(5):943-55. doi: 10.1016/j.cell.2007.03.046. Cell. 2007. PMID: 17540174 Free PMC article. - Free N-linked oligosaccharide chains: formation and degradation.
Suzuki T, Funakoshi Y. Suzuki T, et al. Glycoconj J. 2006 Jul;23(5-6):291-302. doi: 10.1007/s10719-006-6975-x. Glycoconj J. 2006. PMID: 16897173 Review. - Prb1 Protease Activity Is Required for Its Recognition by the F-Box Protein Saf1.
Mark KG, Meza-Gutierrez F, Johnson JR, Newton BW, Krogan NJ, Toczyski DP. Mark KG, et al. Biochemistry. 2015 Jul 28;54(29):4423-6. doi: 10.1021/acs.biochem.5b00504. Epub 2015 Jul 17. Biochemistry. 2015. PMID: 26161950 Free PMC article. - Wild type RTA and less toxic variants have distinct requirements for Png1 for their depurination activity and toxicity in Saccharomyces cerevisiae.
Yan Q, Li XP, Tumer NE. Yan Q, et al. PLoS One. 2014 Dec 1;9(12):e113719. doi: 10.1371/journal.pone.0113719. eCollection 2014. PLoS One. 2014. PMID: 25436896 Free PMC article.
References
- Lyko F, Martoglio B, Jungnickel B, Rapoport T A, Dobberstein B. J Biol Chem. 1995;270:19873–19878. - PubMed
- Moriyama T, Sather S K, McGee T P, Simoni R D. J Biol Chem. 1998;273:22037–22043. - PubMed
- Xiong X, Chong E, Skach W R. J Biol Chem. 1999;274:2616–2624. - PubMed
- Lehner P J, Trowsdale J. Curr Biol. 1998;8:R605–R608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases