The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light - PubMed (original) (raw)
The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light
P E Gibbs et al. Proc Natl Acad Sci U S A. 2000.
Abstract
In Saccharomyces cerevisiae, most mutations induced by a wide range of mutagens arise during translesion replication employing the REV1 gene product and DNA polymerase zeta. As part of an effort to investigate mammalian mutagenic mechanisms, we have identified cDNA clones of the human homologs of the yeast REV genes and examined their function in UV mutagenesis. Previously, we described the isolation of a human homolog of yeast REV3, the catalytic subunit of pol zeta, and here report the identification and sequence of a human homolog of yeast REV1. This gene was isolated by identifying an expressed sequence tag encoding a peptide with similarity to the C terminus of yeast Rev1p, followed by sequencing of the clone and retrieval of the remaining cDNA by 5' rapid amplification of cDNA ends. The human gene encodes an expected protein of 1,251 residues, compared with 985 residues in the yeast protein. The proteins share two amino-terminal regions of approximately 100 residues with 41% and 20% identity, a region of approximately 320 residues with 31% identity, and a central motif in which 11 of 13 residues are identical. Human cells expressing high levels of an hREV1 antisense RNA grew normally, and were not more sensitive to the cytotoxic effect of 254 nm UV radiation than cells lacking antisense RNA. However, the frequencies of 6-thioguanine resistance mutants induced by UV in the cells expressing antisense hREV1 RNA were significantly lower than in the control (P = 0.01), suggesting that the human gene has a function similar to that of the yeast homolog.
Figures
Figure 1
(A) Sequence of the translation product of hREV1 mRNA. The underlined sequences numbered I–IX are possible sequence motifs, and the highlighted residues in motifs I, III, and VI are known to be important for function. (B) Depiction of regions of similarity to C. elegans and D. melanogaster sequences (open boxes) together with the location of the predicted motifs. The darker filled boxes indicate the location of motifs III and VI.
Figure 2
Schematic representation of the alignment of hREV1 and yeast REV1 protein sequences. Regions with significant identity are shaded, and the percent identity is indicated.
Figure 3
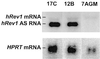
Northern blot analysis of the level of expression of hREV1 antisense RNA in the cell strains. RNA extracted from the two derivative cell strains, 7AGM-17C and 7AGM-12B, which had been transfected with plasmid pR1P27-AS containing the 4,117-bp sequence of the hREV1 RNA in an antisense orientation, and from their nontransfected parental cell strain, MSU-1.2-7AGM, and analyzed for expression of hREV1 antisense RNA (hREV1 AS RNA) and/or the endogenous hREV1 sense RNA. The latter mRNA, which is ≈4.4 kbp in length, can be seen as a faint band just above the antisense band in the first two lanes, and in the third lane. The lower band, approximately 1.4 kbp in length, is the endogenous HPRT mRNA, which was used to normalize the amount of RNA loaded per lane. There was no significant difference between the two derivative cell strains in the level of expression of the antisense RNA.
Figure 4
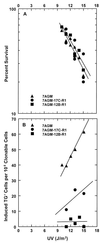
Percent survival (A) and frequencies of UV-induced 6-thioguanine resistant mutants (B) in the parent cell strain 7AGM and the hREV1 antisense RNA-expressing cell strains 7AGM-17C-R1 and 7AGM-12B-R1, plotted against fluence of 254 nm UV. The data are the average of results from four experiments with the parent strain, 7AGM; three experiments with 7AGM-17C-R1 cells; and two experiments with 7AGM-12B-R1cells. The background frequencies of mutants, which have been subtracted to obtain the frequencies induced above the background frequency by UV (23), are cited in the text. The mutant frequencies observed for untreated control and each UV fluence were corrected for the cloning efficiency of the cells determined after an 8-day expression period, i.e., at the time they were plated into selection medium containing 6-thioguanine. The average cloning efficiencies for control and UV-irradiated cells were: 7AGM cells, 33.3% ± 1.4% (SEM for four independent experiments); 7AGM-17C-R1 cells, 26.7 ± 1.3% (SEM for three independent experiments); and 7AGM-12B-R1 cells, 31.8% ± 0.9% (SEM for two independent experiments).
Similar articles
- A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta.
Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. Gibbs PE, et al. Proc Natl Acad Sci U S A. 1998 Jun 9;95(12):6876-80. doi: 10.1073/pnas.95.12.6876. Proc Natl Acad Sci U S A. 1998. PMID: 9618506 Free PMC article. - Alteration of ultraviolet-induced mutagenesis in yeast through molecular modulation of the REV3 and REV7 gene expression.
Rajpal DK, Wu X, Wang Z. Rajpal DK, et al. Mutat Res. 2000 Oct 16;461(2):133-43. doi: 10.1016/s0921-8777(00)00047-1. Mutat Res. 2000. PMID: 11018586 - Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p.
Lawrence CW, Maher VM. Lawrence CW, et al. Philos Trans R Soc Lond B Biol Sci. 2001 Jan 29;356(1405):41-6. doi: 10.1098/rstb.2000.0001. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11205328 Free PMC article. Review. - Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein.
Lawrence CW, Maher VM. Lawrence CW, et al. Biochem Soc Trans. 2001 May;29(Pt 2):187-91. doi: 10.1042/0300-5127:0290187. Biochem Soc Trans. 2001. PMID: 11356151 Review.
Cited by
- Caffeine abolishes the ultraviolet-induced REV3 translesion replication pathway in mouse cells.
Takezawa J, Aiba N, Kajiwara K, Yamada K. Takezawa J, et al. Int J Mol Sci. 2011;12(12):8513-29. doi: 10.3390/ijms12128513. Epub 2011 Nov 29. Int J Mol Sci. 2011. PMID: 22272088 Free PMC article. - Using ultra-sensitive next generation sequencing to dissect DNA damage-induced mutagenesis.
Wang K, Ma X, Zhang X, Wu D, Sun C, Sun Y, Lu X, Wu CI, Guo C, Ruan J. Wang K, et al. Sci Rep. 2016 Apr 28;6:25310. doi: 10.1038/srep25310. Sci Rep. 2016. PMID: 27122023 Free PMC article. - RAD18 and associated proteins are immobilized in nuclear foci in human cells entering S-phase with ultraviolet light-induced damage.
Watson NB, Nelson E, Digman M, Thornburg JA, Alphenaar BW, McGregor WG. Watson NB, et al. Mutat Res. 2008 Dec 15;648(1-2):23-31. doi: 10.1016/j.mrfmmm.2008.09.006. Epub 2008 Sep 24. Mutat Res. 2008. PMID: 18926833 Free PMC article. - Rev1 is a base excision repair enzyme with 5'-deoxyribose phosphate lyase activity.
Prasad R, Poltoratsky V, Hou EW, Wilson SH. Prasad R, et al. Nucleic Acids Res. 2016 Dec 15;44(22):10824-10833. doi: 10.1093/nar/gkw869. Epub 2016 Sep 28. Nucleic Acids Res. 2016. PMID: 27683219 Free PMC article. - The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis.
Jansen JG, Tsaalbi-Shtylik A, Langerak P, Calléja F, Meijers CM, Jacobs H, de Wind N. Jansen JG, et al. Nucleic Acids Res. 2005 Jan 13;33(1):356-65. doi: 10.1093/nar/gki189. Print 2005. Nucleic Acids Res. 2005. PMID: 15653636 Free PMC article.
References
- Lawrence C W, Hinkle D C. Cancer Surv. 1996;28:21–31. - PubMed
- Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. - PubMed
- Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. - PubMed
- Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous