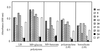Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds - PubMed (original) (raw)
Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds
M Espinosa-Urgel et al. J Bacteriol. 2000 May.
Abstract
Many agricultural uses of bacteria require the establishment of efficient bacterial populations in the rhizosphere, for which colonization of plant seeds often constitutes a critical first step. Pseudomonas putida KT2440 is a strain that colonizes the rhizosphere of a number of agronomically important plants at high population densities. To identify the functions involved in initial seed colonization by P. putida KT2440, we subjected this strain to transposon mutagenesis and screened for mutants defective in attachment to corn seeds. Eight different mutants were isolated and characterized. While all of them showed reduced attachment to seeds, only two had strong defects in their adhesion to abiotic surfaces (glass and different plastics). Sequences of the loci affected in all eight mutants were obtained. None of the isolated genes had previously been described in P. putida, although four of them showed clear similarities with genes of known functions in other organisms. They corresponded to putative surface and membrane proteins, including a calcium-binding protein, a hemolysin, a peptide transporter, and a potential multidrug efflux pump. One other showed limited similarities with surface proteins, while the remaining three presented no obvious similarities with known genes, indicating that this study has disclosed novel functions.
Figures
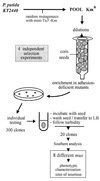
FIG. 1
Summary of the screening strategy used to isolate mus derivatives of P. putida KT2440. Details are presented in the text.
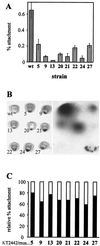
FIG. 2
Adhesion of P. putida KT2440 (wild type [wt]) and mus mutants to corn seeds. (A) Quantitation of attachment of P. putida KT2440 and eight mus mutants to corn seeds. After 1 h of incubation with each bacterial suspension, seeds were washed and disrupted, and the number of attached cells was estimated as CFU after plating serial dilutions. Results are presented as percentage of attached cells with respect to the number of cells inoculated (average of at least three independent experiments). (B) Adhesion to corn seeds (left) of KT2440 and mus mutants harboring plasmid pDLDLUX, visualized by overnight exposure of film (right). (C) Adhesion of mus mutants (white bars) coinoculated with KT2442 (grey bar). Relative percentages of cells of each strain attached to the seeds (average of three independent assays) are shown.
FIG. 3
Biofilm formation on different abiotic surfaces, quantified by staining attached cells with crystal violet and measuring _A_600 after solubilization of the stain with ethanol. Results are the average of four independent assays. wt, wild type.
FIG. 4
Surface proteins of KT2440 and mus strains. Surface proteins were analyzed by SDS-PAGE on a 12% polyacrylamide gel and staining with Coomassie brilliant blue. Bands showing less intensity in the mutants than in the parental strain (wild type [wt]) are marked with black arrows; bands with more intensity in the mutants than in KT2440 are indicated with white arrows. Positions of molecular weight markers are indicated in kilodaltons.
Similar articles
- Role of iron and the TonB system in colonization of corn seeds and roots by Pseudomonas putida KT2440.
Molina MA, Godoy P, Ramos-González MI, Muñoz N, Ramos JL, Espinosa-Urgel M. Molina MA, et al. Environ Microbiol. 2005 Mar;7(3):443-9. doi: 10.1111/j.1462-2920.2005.00720.x. Environ Microbiol. 2005. PMID: 15683404 - A two-partner secretion system is involved in seed and root colonization and iron uptake by Pseudomonas putida KT2440.
Molina MA, Ramos JL, Espinosa-Urgel M. Molina MA, et al. Environ Microbiol. 2006 Apr;8(4):639-47. doi: 10.1111/j.1462-2920.2005.00940.x. Environ Microbiol. 2006. PMID: 16584475 - Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida.
Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. Yousef-Coronado F, et al. FEMS Microbiol Lett. 2008 Nov;288(1):118-24. doi: 10.1111/j.1574-6968.2008.01339.x. FEMS Microbiol Lett. 2008. PMID: 18783437 - Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas.
Duque E, de la Torre J, Bernal P, Molina-Henares MA, Alaminos M, Espinosa-Urgel M, Roca A, Fernández M, de Bentzmann S, Ramos JL. Duque E, et al. Environ Microbiol. 2013 Jan;15(1):36-48. doi: 10.1111/j.1462-2920.2012.02732.x. Epub 2012 Mar 28. Environ Microbiol. 2013. PMID: 22458445 - Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms.
Klausen M, Gjermansen M, Kreft JU, Tolker-Nielsen T. Klausen M, et al. FEMS Microbiol Lett. 2006 Aug;261(1):1-11. doi: 10.1111/j.1574-6968.2006.00280.x. FEMS Microbiol Lett. 2006. PMID: 16842351 Review.
Cited by
- Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482.
Rajewska M, Maciąg T, Narajczyk M, Jafra S. Rajewska M, et al. Int J Mol Sci. 2024 Jul 30;25(15):8351. doi: 10.3390/ijms25158351. Int J Mol Sci. 2024. PMID: 39125921 Free PMC article. - Variability in Maize Seed Bacterization and Survival Correlating with Root Colonization by Pseudomonas Isolates with Plant-Probiotic Traits.
Lorch MG, Valverde C, Agaras BC. Lorch MG, et al. Plants (Basel). 2024 Aug 1;13(15):2130. doi: 10.3390/plants13152130. Plants (Basel). 2024. PMID: 39124248 Free PMC article. - Seedling microbiota engineering using bacterial synthetic community inoculation on seeds.
Arnault G, Marais C, Préveaux A, Briand M, Poisson AS, Sarniguet A, Barret M, Simonin M. Arnault G, et al. FEMS Microbiol Ecol. 2024 Mar 12;100(4):fiae027. doi: 10.1093/femsec/fiae027. FEMS Microbiol Ecol. 2024. PMID: 38503562 Free PMC article. - Role of extracellular matrix components in biofilm formation and adaptation of Pseudomonas ogarae F113 to the rhizosphere environment.
Blanco-Romero E, Garrido-Sanz D, Durán D, Rybtke M, Tolker-Nielsen T, Redondo-Nieto M, Rivilla R, Martín M. Blanco-Romero E, et al. Front Microbiol. 2024 Jan 25;15:1341728. doi: 10.3389/fmicb.2024.1341728. eCollection 2024. Front Microbiol. 2024. PMID: 38333580 Free PMC article. - Root colonization by beneficial rhizobacteria.
Liu Y, Xu Z, Chen L, Xun W, Shu X, Chen Y, Sun X, Wang Z, Ren Y, Shen Q, Zhang R. Liu Y, et al. FEMS Microbiol Rev. 2024 Jan 12;48(1):fuad066. doi: 10.1093/femsre/fuad066. FEMS Microbiol Rev. 2024. PMID: 38093453 Free PMC article.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987.
- Bayliss C, Bent E, Culham D E, MacLellan S, Clarke A J, Brown G L, Wood J M. Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3–canola mutualism: identification of an exudate-inducible sugar transporter. Can J Microbiol. 1997;43:809–818. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases