Distinct protein interaction domains and protein spreading in a complex centromere - PubMed (original) (raw)
. 2000 Apr 1;14(7):783-91.
Affiliations
- PMID: 10766735
- PMCID: PMC316498
Distinct protein interaction domains and protein spreading in a complex centromere
J F Partridge et al. Genes Dev. 2000.
Abstract
Fission yeast (Schizosaccharomyces pombe) centromeres are composed of large (40-100 kb) inverted repeats that display heterochromatic features, thus providing a good model for higher eukaryotic centromeres. The association of three proteins that mediate region-specific silencing across centromere 1 has been mapped by quantitative chromatin immunoprecipitation. Swi6 and Chp1 are confined to the flanking outer repeats and Swi6 can spread across at least 3 kb of extraneous chromatin in cen1. In contrast, Mis6 coats the inner repeats and central core. tRNA genes demarcate this transition zone. These analyses clearly define two distinct domains within this complex centromere which interact with different proteins.
Figures
Figure 1
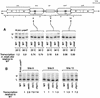
Chp1 and Mis6 silence distinct centromeric domains. (A) mis6-302 specifically alleviates central core silencing, but not silencing of the outer repeats. (B) Chp1, like Clr4, Rik1 and Swi6 is required for silencing of the outer repeats, but not the central core. For both A and B, competitive radioactive PCR was performed on cDNA generated by RT–PCR from strains with ura4+ (U) inserted within the central core (site 9), the inner and outer repeats of centromere 1 (sites 6 and 13) or in euchromatin (R.Int::ura4+) and a fully expressed ura4–DS/E minigene (L) at the ura4 locus. Separated PCR products were quantified. Levels of ura4 (U) were normalized to ura4–DS/E (L) in the mutant strains and expressed relative to values obtained for the wild-type background for each insertion site. In A, mis6-302 and wild-type strains were grown at 25°C, or shifted to the nonpermissive temperature for 4 hr prior to RNA extraction.
Figure 2
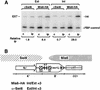
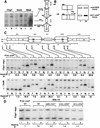
Mis6 and Swi6 associate with distinct domains of cen1. (A) Strains with ura4+ at different positions within cen1 (U) and the ura4–DS/E minigene at the ura4 locus (L), were formaldehyde fixed. Chromatin was prepared and sheared to 500–1000 bp prior to immunoprecipitation with anti-Swi6 or anti-HA antibodies (Mis6–3xHA strains). Recovered immunoprecipitated DNAs (ip) were compared with input crude DNA (c) by competitive PCR of ura4+. (B) Quantitative chromatin immunoprecipitation shows that Swi6 associates with the outer repeats, and Mis6 with the inner repeats and central core of cen1. Chromatin immunoprecipitation from strains with ura4 inserted at different positions within cen1 (represented by cartoon at right) is shown. For some insertion sites, two strains bearing different orientations of ura4+ were tested (open arrowheads; see Allshire et al. 1995). Results for both Swi6 and HA immunoprecipitation of Mis6–3xHA strains is shown. (C) Quantitation of these results is plotted. Levels of enrichment of ura4 (U) at each insertion site were quantified relative to ura4–DS/E (L) and normalized to values obtained for strains bearing a random integrant of ura4 (R.Int::ura4+). Results for several experiments investigating Swi6 and Mis6–3xHA immunoprecipitation are plotted. (D) Swi6 immunoprecipitation of centromeric chromatin is Clr4, Rik1, and Swi6 dependent. Swi6 chromatin immunoprecipitation of ura4+ (U) at centromeric site 13 is abolished in yeast bearing mutations in clr4 and rik1.
Figure 3
tRNA genes demarcate a transition zone between Mis6 and Swi6 cen1 association. (A) PCR primers were designed to amplify sequences internal (Int) or external (Ext), indicated by the hatched boxes in imr1L, surrounding the tRNAAla and tRNAGlu genes (filled arrowheads, B) from immunoprecipitated samples. Swi6 and Mis6–3xHA immunoprecipitates were assessed for the levels of Ext and Int PCR products relative to the fbp1+ control. (B) Diagram of cen1 showing the mapping of the transition zone between centromeric Mis6–3xHA and Swi6 to a region containing tRNA genes. Mis6–3xHA immunoprecipitates gave an average threefold enrichment of Int product over Ext, and Swi6, a threefold enrichment of Ext over Int.
Figure 4
Chp1 displays a similar cen1 association pattern as Swi6. (A) Multiplex PCR was performed to detect association of Chp1–6xMyc with centromeric chromatin. Primers were designed to two sites (imr/otr junction and a region of cnt1), which give amplification specifically from cen1 sequences, and to the euchromatic fbp1+ gene locus to act as a control for nonspecific association. Chp1–6xMyc and Swi6 immunoprecipitates both showed enrichment of the imr/otr product relative to fbp1 and showed no enrichment for the central core sequence (cnt1). In contrast, Mis6–3xHA immunoprecipitates showed enrichment for cnt1 and not for imr/otr. (B) Chp1–6xMyc interactions across cen1 were mapped by specific PCR from immunoprecipitates of strains with different cen1–ura4 insertions using various primers from cen1 and one primer anchored in the ura4+ gene. Enrichment of centromeric ura4+ by immunoprecipitation is reflected by increased intensity of the smaller PCR products, which vary in size from different strains, depending on the location of the centromeric primers, relative to the large PCR product of constant size that reflects association with the euchromatic ura4–DS/E locus. (C) Using this assay, Chp1–6xMyc and Swi6 associate with the flanking repeats but not the central core of cen1. Relative ip values are an average of 2 (Swi6), and 3 (Chp1–myc) experiments. (D) Chp1–6xMyc immunoprecipitation at site 13 is dependent on Clr4 and Rik1 but not Swi6.
Figure 5
Swi6 can spread over noncentromeric DNA. Strains with 1.3 kb or 3 kb surrounding ura4+ and inserted at site 13 showed equivalent levels of Swi6 chromatin immunoprecipitation of ura4+ as strains with only ura4+ at that site.
Similar articles
- Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells.
Appelgren H, Kniola B, Ekwall K. Appelgren H, et al. J Cell Sci. 2003 Oct 1;116(Pt 19):4035-42. doi: 10.1242/jcs.00707. Epub 2003 Aug 19. J Cell Sci. 2003. PMID: 12928332 - Requirement of heterochromatin for cohesion at centromeres.
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Bernard P, et al. Science. 2001 Dec 21;294(5551):2539-42. doi: 10.1126/science.1064027. Epub 2001 Oct 11. Science. 2001. PMID: 11598266 - Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast.
Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Nonaka N, et al. Nat Cell Biol. 2002 Jan;4(1):89-93. doi: 10.1038/ncb739. Nat Cell Biol. 2002. PMID: 11780129 - Centromere structure and function in budding and fission yeasts.
Carbon J, Clarke L. Carbon J, et al. New Biol. 1990 Jan;2(1):10-9. New Biol. 1990. PMID: 2078550 Review. - Studies on the mechanism of RNAi-dependent heterochromatin assembly.
Moazed D, Bühler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villén J, Gygi SP. Moazed D, et al. Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
- Myb-domain protein Teb1 controls histone levels and centromere assembly in fission yeast.
Valente LP, Dehé PM, Klutstein M, Aligianni S, Watt S, Bähler J, Cooper JP. Valente LP, et al. EMBO J. 2013 Feb 6;32(3):450-60. doi: 10.1038/emboj.2012.339. Epub 2013 Jan 11. EMBO J. 2013. PMID: 23314747 Free PMC article. - Histone modifications within the human X centromere region.
Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA. Mravinac B, et al. PLoS One. 2009 Aug 12;4(8):e6602. doi: 10.1371/journal.pone.0006602. PLoS One. 2009. PMID: 19672304 Free PMC article. - The contradictory definitions of heterochromatin: transcription and silencing.
Huisinga KL, Brower-Toland B, Elgin SC. Huisinga KL, et al. Chromosoma. 2006 Apr;115(2):110-22. doi: 10.1007/s00412-006-0052-x. Epub 2006 Feb 28. Chromosoma. 2006. PMID: 16506022 Review. - The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association.
Kerres A, Vietmeier-Decker C, Ortiz J, Karig I, Beuter C, Hegemann J, Lechner J, Fleig U. Kerres A, et al. Mol Biol Cell. 2004 Dec;15(12):5255-67. doi: 10.1091/mbc.e04-06-0443. Epub 2004 Sep 15. Mol Biol Cell. 2004. PMID: 15371542 Free PMC article. - A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere.
Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH. Lo AW, et al. EMBO J. 2001 Apr 17;20(8):2087-96. doi: 10.1093/emboj/20.8.2087. EMBO J. 2001. PMID: 11296241 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. - PMC - PubMed
- Allshire RC. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. Transcriptional silencing in the fission yeast: A manifestation of higher order chromosome structure and functions; pp. 443–466.
- Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
- Allshire RC, Nimmo ER, Ekwall K, Javerzat J-P, Cranston G. Mutations derepressing silent domains within fission yeast centromeres disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases