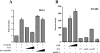The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression - PubMed (original) (raw)
The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression
K I Takemaru et al. J Cell Biol. 2000.
Abstract
Beta-catenin plays a pivotal role in the transcriptional activation of Wnt-responsive genes by binding to TCF/LEF transcription factors. Although it has been suggested that the COOH-terminal region of beta-catenin functions as an activation domain, the mechanisms of activation remain unclear. To screen for potential transcriptional coactivators that bind to the COOH-terminal region of beta-catenin, we used a novel yeast two-hybrid system, the Ras recruitment system (RRS) that detects protein-protein interactions at the inner surface of the plasma membrane. Using this system, we isolated the CREB-binding protein (CBP). Armadillo (Arm) repeat 10 to the COOH terminus of beta-catenin is involved in binding to CBP, whereas beta-catenin interacts directly with the CREB-binding domain of CBP. Beta-catenin synergizes with CBP to stimulate the activity of a synthetic reporter in vivo. Conversely, beta-catenin-dependent transcriptional activation is repressed by E1A, an antagonist of CBP function, but not by an E1A mutant that does not bind to CBP. The activation of Wnt target genes such as siamois and Xnr3 in Xenopus embryos is also sensitive to E1A. These findings suggest that CBP provides a link between beta-catenin and the transcriptional machinery, and possibly mediates the oncogenic function of beta-catenin.
Figures
Figure 1
Interaction between β-catenin and CBP. A, Complementation of the cdc25-2 mutation through the interaction of β-catenin COOH-terminal region with CBP. The temperature-sensitive cdc25-2 yeast cells were cotransformed with the indicated plasmids and the CBP expression plasmid isolated in the screening, and incubated on galactose containing plates either at 25 (left) or 37°C (right). B, Fine mapping of the β-catenin domain that binds to CBP in yeast. Cells were cotransformed with the indicated plasmids together with the CBP expression plasmid, and tested for the growth at 37°C as described for A. The numbers indicate the locations in the amino acid sequence of β-catenin. ++, Efficient growth; +, poor growth; −, no growth. C, β-Catenin directly binds to CREB-binding domain of CBP. Bacterially expressed and purified His-tagged βcatR10-C or _myc-_tagged β-catenin was incubated with buffer (lane 5), GST (lanes 2 and 6), or GST-CBP 451–682 (lanes 3 and 7). After extensive washing, bound proteins were eluted and resolved by SDS-PAGE. βCatR10-C and β-catenin were detected by an anti-His antibody and an anti-myc antibody, respectively. Approximately 10% of the input is shown (lanes 1 and 4). D, Coimmunoprecipitation of CBP and β-catenin. COS7 cells were transfected with expression plasmids for CBP and/or _myc_-tagged pt β-catenin as indicated. Cell lysates were immunoprecipitated with anti-CBP polyclonal antibodies 2 d after transfection. The immunoprecipitates were separated by SDS-PAGE and subjected to Western blotting with anti-myc antibodies.
Figure 2
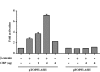
CBP synergizes with β-catenin to activate transcription. HeLa cells were transiently transfected with 1 μg of pTOPFLASH or pFOPFLASH reporter, indicated amounts of CBP expression plasmid, either with or without 1 μg of pt β-catenin–encoding plasmid. Transfections were carried out in triplicate and the means ± SD are shown. Where they are not evident, this is due to minimal variability between transfections.
Figure 3
E1A represses β-catenin–mediated transcriptional activation. A, The reporter plasmid pTOPFLASH (1 μg) was transfected into HeLa cells with or without 1 μg of pt β-catenin–encoding plasmid. The expression vector encoding E1A or E1A mutCBP (100 or 500 ng) were cotransfected as indicated. B, SW480 cells were cotransfected with 100 ng of the pTOPFLASH reporter, and increasing amounts (100 or 400 ng) of CBP expression plasmid, 5 ng each of the plasmids encoding E1A WT, E1A mutRB, or E1A mutCBP, or 50 ng of dnLEF-1 expression plasmid as indicated. The activity of the pTOPFLASH reporter in the absence of exogenous CBP, E1A, or dnLEF-1 was set at 100%.
Figure 4
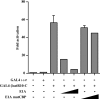
Endogenous CBP is required for transactivation by GAL4-βcatR10-C fusion protein. HeLa cells were transfected with 1 μg of GAL4 reporter construct, pFR-Luc, 50 ng of GAL4 1–147 or GAL4-βcatR10-C, and increasing amounts of E1A or E1A mutCBP expression plasmid (25 or 500 ng) in combinations as indicated.
Figure 5
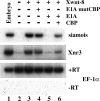
The activation of endogenous Wnt-responsive genes is sensitive to E1A. Both blastomeres of two-cell stage Xenopus embryos were injected with 20 pg of Xwnt-8, 0.5 ng of E1A mutCBP, or E1A, and 3.2 ng of CBP RNAs as indicated. Animal cap explants were dissected at stage 8 to 9 and analyzed for gene expression by RT-PCR.
Similar articles
- Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and beta-catenin.
Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. Labalette C, et al. Mol Cell Biol. 2004 Dec;24(24):10689-702. doi: 10.1128/MCB.24.24.10689-10702.2004. Mol Cell Biol. 2004. PMID: 15572674 Free PMC article. - The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates.
Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. Hecht A, et al. EMBO J. 2000 Apr 17;19(8):1839-50. doi: 10.1093/emboj/19.8.1839. EMBO J. 2000. PMID: 10775268 Free PMC article. - Nuclear localization of Duplin, a beta-catenin-binding protein, is essential for its inhibitory activity on the Wnt signaling pathway.
Kobayashi M, Kishida S, Fukui A, Michiue T, Miyamoto Y, Okamoto T, Yoneda Y, Asashima M, Kikuchi A. Kobayashi M, et al. J Biol Chem. 2002 Feb 22;277(8):5816-22. doi: 10.1074/jbc.M108433200. Epub 2001 Dec 13. J Biol Chem. 2002. PMID: 11744694 - TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling.
Brantjes H, Barker N, van Es J, Clevers H. Brantjes H, et al. Biol Chem. 2002 Feb;383(2):255-61. doi: 10.1515/BC.2002.027. Biol Chem. 2002. PMID: 11934263 Review. - Taking the road less traveled - the therapeutic potential of CBP/β-catenin antagonists.
Kahn M. Kahn M. Expert Opin Ther Targets. 2021 Sep;25(9):701-719. doi: 10.1080/14728222.2021.1992386. Epub 2021 Oct 27. Expert Opin Ther Targets. 2021. PMID: 34633266 Free PMC article. Review.
Cited by
- Wnt signaling: the β-cat(enin)'s meow.
Bauer M, Willert K. Bauer M, et al. Genes Dev. 2012 Jan 15;26(2):105-9. doi: 10.1101/gad.185280.111. Genes Dev. 2012. PMID: 22279043 Free PMC article. - Frequent amplification of AIB1, a critical oncogene modulating major signaling pathways, is associated with poor survival in gastric cancer.
Shi J, Liu W, Sui F, Lu R, He Q, Yang Q, Lv H, Shi B, Hou P. Shi J, et al. Oncotarget. 2015 Jun 10;6(16):14344-59. doi: 10.18632/oncotarget.3852. Oncotarget. 2015. PMID: 25970779 Free PMC article. - Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors.
Wöhrle S, Wallmen B, Hecht A. Wöhrle S, et al. Mol Cell Biol. 2007 Dec;27(23):8164-77. doi: 10.1128/MCB.00555-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923689 Free PMC article. - The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation.
Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. Barker N, et al. EMBO J. 2001 Sep 3;20(17):4935-43. doi: 10.1093/emboj/20.17.4935. EMBO J. 2001. PMID: 11532957 Free PMC article. - CBP/p300 are bimodal regulators of Wnt signaling.
Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. Li J, et al. EMBO J. 2007 May 2;26(9):2284-94. doi: 10.1038/sj.emboj.7601667. Epub 2007 Apr 5. EMBO J. 2007. PMID: 17410209 Free PMC article.
References
- Barker N., Morin P.J., Clevers H. The Yin–Yang of TCF/β-catenin signaling. Adv. Cancer Res. 2000;77:1–24. - PubMed
- Behrens J., von Kries J.P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
- Brannon M., Brown J.D., Bates R., Kimelman D., Moon R.T. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous