Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol - PubMed (original) (raw)
Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol
G F Bammert et al. Antimicrob Agents Chemother. 2000 May.
Abstract
Enzymes in the ergosterol-biosynthetic pathway are the targets of a number of antifungal agents including azoles, allylamines, and morpholines. In order to understand the response of Saccharomyces cerevisiae to perturbations in the ergosterol pathway, genome-wide transcript profiles following exposure to a number of antifungal agents targeting ergosterol biosynthesis (clotrimazole, fluconazole, itraconazole, ketoconazole, voriconazole, terbinafine, and amorolfine) were obtained. These profiles were compared to the transcript profiles of strains containing deletions of one of the late-stage ergosterol genes: ERG2, ERG5, or ERG6. A total of 234 genes were identified as responsive, including the majority of genes from the ergosterol pathway. Expression of several responsive genes, including ERG25, YER067W, and YNL300W, was also monitored by PCR over time following exposure to ketoconazole. The kinetics of transcriptional response support the conditions selected for the microarray experiment. In addition to ergosterol-biosynthetic genes, 36 mitochondrial genes and a number of other genes with roles related to ergosterol function were responsive, as were a number of genes responsive to oxidative stress. Transcriptional changes related to heme biosynthesis were observed in cells treated with chemical agents, suggesting an additional effect of exposure to these compounds. The expression profile in response to a novel imidazole, PNU-144248E, was also determined. The concordance of responsive genes suggests that this compound has the same mode of action as other azoles. Thus, genome-wide transcript profiles can be used to predict the mode of action of a chemical agent as well as to characterize expression changes in response to perturbation of a metabolic pathway.
Figures
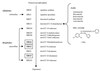
FIG. 1
Treatments used in the study and their relationships to ergosterol biosynthesis. Gene names are as listed in reference 9). Genes in deletion strains are boldfaced and boxed. Arrows point to the sites of action of the antifungal agents. The structure of PNU-144248E is shown under the azoles.
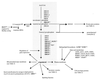
FIG. 2
Genes involved in the biosynthesis of ergosterol and membrane components. Boldfaced genes were responsive in the study; boldface italics indicate genes with decreased transcript levels. Superscripts indicate the number of treatments to which the gene responded. The first number in each superscript is the number of genetic perturbations (out of a total of three) which elicited a response, and the second number is the number of drug treatments (out of a total of eight) which elicited a response. Lists of genes were obtained from the YPD (18) and reference .
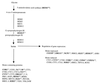
FIG. 3
Genes involved in the biosynthesis and utilization of heme. Boldface, italics, and superscripts are as described for Fig. 2. Lists of genes were obtained from the YPD (18).
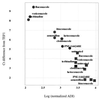
FIG. 4
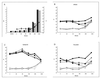
Comparison of expression measurements by RT-PCR and microarrays. Each data point represents the measurement of a given gene by each method under a specific treatment, as indicated by the label. Squares, ERG25; circles, YER067W; solid symbols, untreated control. The vertical axis is the change from the TEF1 level; larger numbers signify more cycles needed and thus less starting material in the sample. The horizontal axis indicates the microarray intensity in ADI units, normalized as described in Materials and Methods; ADI units are proportional to transcript levels.
FIG. 5
Expression time course following exposure to ketoconazole. (A) Culture OD at the times sampled (solid bars, untreated culture; heavily shaded bars, 4 μM ketoconazole; open bars, 8 μM ketoconazole; light shaded bars, 16 μM ketoconazole) and change in TEF1 level as culture goes into saturation (line, average of three measurements of untreated culture in panels B, C, and D; error bars, standard errors). (B through D) Responses of ERG25, YER067W, and YNL300W transcripts (solid symbols) and an internal TEF1 control (open symbols). Culture was left untreated (circles) or treated with ketoconazole at 4 μM (triangles), 8 μM (squares), or 16 μM (diamonds).
Similar articles
- Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces cerevisiae.
Bhattacharya S, Esquivel BD, White TC. Bhattacharya S, et al. mBio. 2018 Jul 24;9(4):e01291-18. doi: 10.1128/mBio.01291-18. mBio. 2018. PMID: 30042199 Free PMC article. - Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae.
Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. Agarwal AK, et al. J Biol Chem. 2003 Sep 12;278(37):34998-5015. doi: 10.1074/jbc.M306291200. Epub 2003 Jun 24. J Biol Chem. 2003. PMID: 12824174 - Coordinated Regulation of Membrane Homeostasis and Drug Accumulation by Novel Kinase STK-17 in Response to Antifungal Azole Treatment.
Hu C, Zhou M, Cao X, Xue W, Zhang Z, Li S, Sun X. Hu C, et al. Microbiol Spectr. 2022 Feb 23;10(1):e0012722. doi: 10.1128/spectrum.00127-22. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196787 Free PMC article. - The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn.
Lv QZ, Yan L, Jiang YY. Lv QZ, et al. Virulence. 2016 Aug 17;7(6):649-59. doi: 10.1080/21505594.2016.1188236. Epub 2016 May 24. Virulence. 2016. PMID: 27221657 Free PMC article. Review. - Ergosterol biosynthesis inhibition: a target for antifungal agents.
Barrett-Bee K, Dixon G. Barrett-Bee K, et al. Acta Biochim Pol. 1995;42(4):465-79. Acta Biochim Pol. 1995. PMID: 8852337 Review.
Cited by
- Novel yeast-based biosensor for environmental monitoring of tebuconazole.
Mendes F, Miranda E, Amaral L, Carvalho C, Castro BB, Sousa MJ, Chaves SR. Mendes F, et al. Appl Microbiol Biotechnol. 2024 Dec;108(1):10. doi: 10.1007/s00253-023-12944-z. Epub 2024 Jan 3. Appl Microbiol Biotechnol. 2024. PMID: 38170307 Free PMC article. - Potential of Alpha-(α)-Solanine as a Natural Inhibitor of Fungus Causing Leaf Spot Disease in Strawberry.
Xu N, Lu H, Yi X, Peng S, Huang X, Zhang Y, He C. Xu N, et al. Life (Basel). 2023 Feb 6;13(2):450. doi: 10.3390/life13020450. Life (Basel). 2023. PMID: 36836807 Free PMC article. - Genome-Wide Identification of Cellular Pathways and Key Genes That Respond to Sodium Bicarbonate Stress in Saccharomyces cerevisiae.
Cao X, An T, Fu W, Zhang J, Zhao H, Li D, Jin X, Liu B. Cao X, et al. Front Microbiol. 2022 Apr 12;13:831973. doi: 10.3389/fmicb.2022.831973. eCollection 2022. Front Microbiol. 2022. PMID: 35495664 Free PMC article. - Clotrimazole-Induced Oxidative Stress Triggers Novel Yeast Pkc1-Independent Cell Wall Integrity MAPK Pathway Circuitry.
Sellers-Moya Á, Nuévalos M, Molina M, Martín H. Sellers-Moya Á, et al. J Fungi (Basel). 2021 Aug 9;7(8):647. doi: 10.3390/jof7080647. J Fungi (Basel). 2021. PMID: 34436186 Free PMC article. - Modulation of ERG gene expression in fluconazole-resistant human and animal isolates of Trichophyton verrucosum.
Gnat S, Łagowski D, Dyląg M, Ptaszyńska A, Nowakiewicz A. Gnat S, et al. Braz J Microbiol. 2021 Dec;52(4):2439-2446. doi: 10.1007/s42770-021-00585-1. Epub 2021 Aug 5. Braz J Microbiol. 2021. PMID: 34351602 Free PMC article.
References
- Amillet J-M, Buisson N, Labbe-Bois R. Characterization of an upstream activation sequence and two Rox1p-responsive sites controlling the induction of the yeast HEM13 gene by oxygen and heme deficiency. J Biol Chem. 1996;271:24425–24432. - PubMed
- Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. - PubMed
- Boumans H, van Gaalen M C, Grivell L A, Berden J A. Differential inhibition of the yeast bc1 complex by phenanthrolines and ferroin. Implications for structure and catalytic mechanism. J Biol Chem. 1997;272:16753–16760. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases