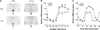Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau - PubMed (original) (raw)
Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau
P L Lowrey et al. Science. 2000.
Abstract
The tau mutation is a semidominant autosomal allele that dramatically shortens period length of circadian rhythms in Syrian hamsters. We report the molecular identification of the tau locus using genetically directed representational difference analysis to define a region of conserved synteny in hamsters with both the mouse and human genomes. The tau locus is encoded by casein kinase I epsilon (CKIepsilon), a homolog of the Drosophila circadian gene double-time. In vitro expression and functional studies of wild-type and tau mutant CKIepsilon enzyme reveal that the mutant enzyme has a markedly reduced maximal velocity and autophosphorylation state. In addition, in vitro CKIepsilon can interact with mammalian PERIOD proteins, and the mutant enzyme is deficient in its ability to phosphorylate PERIOD. We conclude that tau is an allele of hamster CKIepsilon and propose a mechanism by which the mutation leads to the observed aberrant circadian phenotype in mutant animals.
Figures
Fig. 1
(A) The Syrian hamster cross used to map the tau mutation (60). Wild-type males of the outbred M70 strain were mated with outbred female Lak:LVG(Syr) hamsters homozygous for the tau mutation. Siblings of the F1 generation were intercrossed to produce 237 F2 progeny. (B) Representative locomotor activity records of the three circadian phenotypes in the F2 hamster progeny (61). Records are double-plotted so that 48 hours are shown for each horizontal trace. Dark regions represent activity. For the first 7 days, animals were housed in a cycle of 14 hours light, 10 hours dark (LD14 :10), noted by the bar above each record. Animals were transferred to constant darkness (DD) for a total of 3 weeks beginning on day 8 (indicated by a line in the right margin). Top record, wild-type animal with a period of 24 hours; middle record, animal heterozygous for the tau mutation with a period of 22 hours; bottom record, homozygous tau animal with a period of 20 hours. (C) Distribution of period among 237 F2 offspring tested for circadian phenotype.
Fig. 2
(A) Ethidium bromide staining of products from three rounds of GDRDA. Two products (650 and 750 bp) from round three exhibited linkage to the tau mutation (61). (B) Southern hybridization analysis of DNA from the 30 F2 animals comprising the tester and driver pools for GDRDA analysis and the parental animals (61). Œ, the single animal in which a recombination event occurred. The regions of the blot containing the 750-bp signal are shown again below each section with an adjustment in brightness to aid visualization of the signal. Numbers identify individual progeny.
Fig. 3
(A) Genetic map of the Syrian hamster tau locus, showing regions of conserved synteny in mouse and human (69, 76). Genes are as follows: Myoglobin, Mb; aldosterone synthase, Cyp11b2; calcium channel, voltage-dependent, γ subunit 2, Cacng2, stargazer; casein kinase Iɛ, CKIɛ (tau); cadherin EGF LAG seven-pass G-type receptor, Celsr1; elastase, Ela1. A single mouse SSLP marker, D15Mit146, was mapped in hamster (75). In mouse, the locus amplified by the SSR2 hamster primers has been designated D15Nwu1 in the Mouse Genome Database. (B) Electropherogram of sequence around tau mutation region from wild-type and tau homozygous mutant CKIɛ cDNA. An asterisk denotes the point mutation (C→T transition) in the tau cDNA sequence. Amino acid residues are listed under their corresponding nucleotide codons. (C) Alignment of casein kinase I isoforms from various species (100). Dots indicate identities. Kinase subdomains are bounded by the dotted-lines symbol and are numbered with Roman numerals (101). Boxed residues indicate the catalytic loop, the L-9D loop, and the putative nuclear localization signal (NLS). The star symbol marks each of the three residues (Arg178, Gly215, Lys224) that form the anion binding motif. Putative autophosphorylation sites on the carboxyl-terminal tail are indicated by ♦ (82). Drosophiladouble-time short and long-period mutation sites and the hamster tau mutation site are labeled (39, 40). The amino acid sequences of all proteins except hamster and human CKIɛ are omitted following residue 300 because of reduced homology thereafter. Names and GenBank accession numbers are as follows: Mesocricetus auratus CKIɛ (AF242536); Homo sapiens CKIɛ (L37043); Rattus norvegicus CKI8 (L07578); Drosophila melanogaster double-time (AF055583); Arabidopsis thaliana dual specificity kinase (U48779); Saccharomyces cerevisiae HRR25 (M68605).
Fig. 4
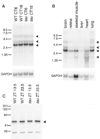
(A) Northern analysis (61) of total RNA prepared from brains of wild-type and homozygous tau hamsters collected at two circadian times, CT6 and CT18. A major transcript of 2.7 kb and minor transcripts of 3.5 and 5.9 kb are indicated by triangles. The blot was normalized by hybridization with probe to mouse GAPDH. (B) Northern analysis (61) of total RNA from wild-type hamster brain, retina, skeletal muscle, liver, heart, and lung tissue. A major transcript of 2.7 kb and minor transcripts of 1.6, 3.7, and 5.9 kb are indicated by triangles. The blot was normalized by hybridization with a probe to mouse GAPDH. (C) Immuno-blot of wild-type and tau hamster hypothalamic tissue extracts (61). Tissues were collected at two zeitgeber (ZT) times (13.5 and 23.5). Blots were probed with antibody to human CKIɛ. A strong signal is evident at ∼48 kD (triangle), the reported size of human CKIɛ (77).
Fig. 5
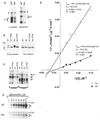
In vitro kinase assays with recombinant homozygous wild type and tau mutant CKIɛ protein. Assays were performed as described (80, 81). Triangles, autophosphorylation products. C and P denote a-casein and phosvitin phosphorylation products, respectively. These results are representative of five independent experiments. (B) Western blot analysis of homozygous wild-type and tau mutant recombinant CKIɛ protein (61). To examine autophosphorylation, kinase assays were performed in the absence of substrate. Following the assay, half of the product was treated with protein phosphatase 2B (PP2B). Blots were incubated with antibody to the S•Tag sequence at the NH2-terminus of the recombinant protein (left) and antibody to the COOH-terminus of human CKIɛ (right). These results are representative of three independent experiments. (C) In vitro kinase assays with homozygous wild type and tau mutant recombinant CKIɛ (84). Enzymes were incubated alone in a kinase reaction prior to addition of substrates to allow autophosphorylation. Half of each reaction was then treated with PP2B. a-casein and phosvitin were used as substrates. C and P denote a-casein and phosvitin phosphorylation products, respectively. The results here are representative of three independent experiments. (D) Kinetic analyses of the kinase activity of homozygous wild-type and tau mutant CKIɛ recombinant protein (61), performed by varying the phosvitin (P) concentration. These results are representative of two independent experiments, each performed in duplicate. (E) The apparent Km and V max values for both enzymes were determined by nonlinear least-squares regression of the untransformed kinetic data (61). The data are represented here graphically on a double-reciprocal plot. Each point represents the average of duplicate samples.
Fig. 6. Figure 6
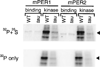
Mutant CKIε is deficient in PERIOD phosphorylation (87). S-agarose beads containing either recombinant wild-type or tau mutant CKIε enzyme were incubated with in vitro– translated, 35S-methionine-labeled substrate and washed extensively. Binding and kinase reactions are indicated for both substrates. Following exposure to detect both the 35S and 32P signals, dried gels were covered with two layers of exposed X-OMAT film to block the 35S signal. PER signal is indicated by filled triangles.
Fig. 7
(A) Effect of the tau mutation on hamster Per1 gene expression in the suprachiasmatic nucleus (61, 102). Example sections showing hamster Per1 in situ hybridization signals in the SCN region. Arrows indicate the position of the SCN. (B) Circadian rhythm of hamster Per1 mRNA expression in the SCN. Samples were collected every 3 circadian hours following 7 days in constant darkness. Onset of locomotor activity for each hamster (CT12; by convention) was used to determine the circadian phase of the sample. Thus, for the tau mutant hamsters, which have an average period length of 20 hours, samples were collected approximately every 2.5 hours. In situ hybridization Per1 signal was quantified as described in (20). Per1 values are plotted relative to the 24-hour average for the wild-type animals. Two-way analysis of variance was significant for circadian time (F-ratio =30.5, P<10-6,df =7), genotype (F-ratio =12.2, _P_=0.0015, df =1) and their interaction (F-ratio =2.52, _P_=0.036, df =7). *Asterisks indicate differences between wild type and tau mutant values at each time point (Tukey’s post hoc, P<0.05). (C) Hamster Per1 mRNA results from (B) plotted in real time. Some points are double-plotted for clarity.
Fig. 8
(A) Cki1 ribbon diagram showing the site of the tau mutation (R183 = hR178) (61). (B) Model of the CKIɛ catalytic surface (61). Coordinates from S. pombe Cki1 are used here to model the analogous region in hamster CKIɛ. ATP (blue) is shown bound in the catalytic cleft. The three residues (yellow) forming the anion binding site are shown binding a sulfate ion (green). In red is the catalytic loop corresponding to residues 128 to 133 (DVKPDN) in hamster CKIɛ (Fig. 3C).
Fig. 9
Model of proposed CKIε role in the mammalian circadian clock, as described in the text. Normal CKIε activity is proposed to cause a delay in the negative feedback signal. LRE, light-responsive element. Blank boxes indicate presumed E-box elements.
Similar articles
- Casein kinase I epsilon gene transfer into the suprachiasmatic nucleus via electroporation lengthens circadian periods of tau mutant hamsters.
Wang H, Ko CH, Koletar MM, Ralph MR, Yeomans J. Wang H, et al. Eur J Neurosci. 2007 Jun;25(11):3359-66. doi: 10.1111/j.1460-9568.2007.05545.x. Eur J Neurosci. 2007. PMID: 17553004 - Casein kinase I: another cog in the circadian clockworks.
Eide EJ, Virshup DM. Eide EJ, et al. Chronobiol Int. 2001 May;18(3):389-98. doi: 10.1081/cbi-100103963. Chronobiol Int. 2001. PMID: 11475410 Review. - Circadian rhythms. Marking time for a kingdom.
Young MW. Young MW. Science. 2000 Apr 21;288(5465):451-3. doi: 10.1126/science.288.5465.451. Science. 2000. PMID: 10798982 No abstract available. - Vasopressin immunoreactivity and release in the suprachiasmatic nucleus of wild-type and tau mutant Syrian hamsters.
Van der Zee EA, Oklejewicz M, Jansen K, Daan S, Gerkema MP. Van der Zee EA, et al. Brain Res. 2002 May 17;936(1-2):38-46. doi: 10.1016/s0006-8993(02)02497-6. Brain Res. 2002. PMID: 11988228 - The biology of the circadian Ck1epsilon tau mutation in mice and Syrian hamsters: a tale of two species.
Loudon AS, Meng QJ, Maywood ES, Bechtold DA, Boot-Handford RP, Hastings MH. Loudon AS, et al. Cold Spring Harb Symp Quant Biol. 2007;72:261-71. doi: 10.1101/sqb.2007.72.073. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18522517 Review.
Cited by
- Breeding and hibernation of captive meadow jumping mice (Zapus hudsonius).
Brem EA, McNulty AD, Israelsen WJ. Brem EA, et al. PLoS One. 2021 May 10;16(5):e0240706. doi: 10.1371/journal.pone.0240706. eCollection 2021. PLoS One. 2021. PMID: 33970917 Free PMC article. - Multi-tissue transcriptional changes and core circadian clock disruption following intensive care.
Hollis HC, Francis JN, Anafi RC. Hollis HC, et al. Front Physiol. 2022 Aug 15;13:942704. doi: 10.3389/fphys.2022.942704. eCollection 2022. Front Physiol. 2022. PMID: 36045754 Free PMC article. - Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes.
Etchegaray JP, Mostoslavsky R. Etchegaray JP, et al. Mol Cell. 2016 Jun 2;62(5):695-711. doi: 10.1016/j.molcel.2016.05.029. Mol Cell. 2016. PMID: 27259202 Free PMC article. Review. - AMP-activated protein kinase as a key molecular link between metabolism and clockwork.
Lee Y, Kim EK. Lee Y, et al. Exp Mol Med. 2013 Jul 26;45(7):e33. doi: 10.1038/emm.2013.65. Exp Mol Med. 2013. PMID: 23887727 Free PMC article. Review.
References
- Aschoff J. Handbook of Behavioral Neurobiology: Biological Rhythms. New York: Plenum; 1981.
- Pittendrigh CS. Annu. Rev. Physiol. 1993;55:16. - PubMed
- Kondo T, Ishiura M. Bioessays. 2000;22:10. - PubMed
- Johnson CH, Golden SS. Annu. Rev. Microbiol. 1999;53:389. - PubMed
- Anderson SL, Kay SA. Adv. Genet. 1997;35:1. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases